
| Home | Syllabus | Schedule | Lecture Notes | Extras | Glossary |

| Home | Syllabus | Schedule | Lecture Notes | Extras | Glossary |
We began by reviewing our introduction to levels of regulation in bacterial gene expression from last time. Cells can regulate transcription or translation, or regulate proteins post-translationally. The fastest response will be post-translational and the slowest will be transcriptional. In terms of resource usage, post-translational regulation is the most expensive, while the rgulation of transcription is the cheapest.
We also reviewed the concepts of positive and negative regulation of transcription. In positive regulation of transcription, the binding of an activator protein to the regulatory region of a gene or operon increases transcription. In negative regulation, the binding of a repressor protein to the regulatory region of a gene or operon decreases transcription.
There are examples of both positive and negative regulation of transcription in the lac operon. The CAP protein, when bound to cAMP, is a positive regulator of the lac operon and other operons. The lac repressor, when not bound to the inducer, is a negative regulator of the lac operon.
We reviewed the circuitry of the lac operon in response to three different states: abundant glucose but no lactose, both glucose and lactose, and lactose alone. These are presented below.
As shown below, in the presence of abundant glucose but no lactose, cAMP levels will be low, so CAP protein will not bind to the lac operon to enhance transcription. In the absence of lactose, the lac repressor will remain bound to the lac operator, preventing transcription.

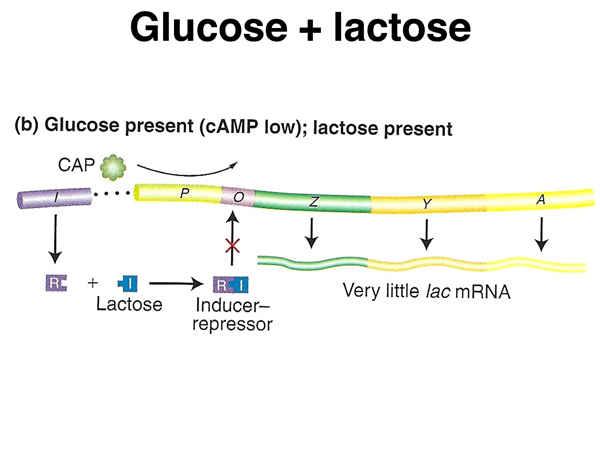
As shown below, in the presence of abundant glucose and also lactose, cAMP levels will be low, so CAP protein will not bind to the lac operon to enhance transcription. In the presence of lactose, the lac repressor will not bind to the lac operator, allowing a low level of transcription.
As shown below, in the absence of glucose and the presence of lactose, cAMP levels will be high, so CAP protein and cAMP will bind to the lac operon to enhance transcription. In the presence of lactose, the lac repressor will not bind to the lac operator, allowing transcription. The presence of CAP-cAMP will cause abundant transcription.

The genetic evidence for our model of the lac operon from last time can be summarized in three tables and accompanying cartoons, shown below.
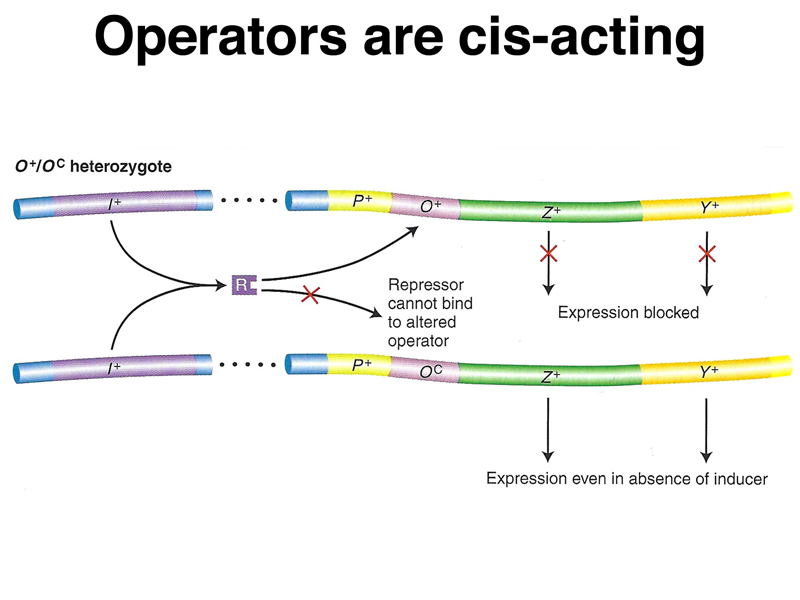
The table below shows the synthesis of β-galactosidase (lacZ) and permease (lacY) in the presence and absence of the inducer IPTG. The data presented in the table are focused on a specific mutation in the operator called an operator-constitutive (lacOC).
| β-galactosidase (lacZ) | permease (lacY) | ||||
| Genotype | - IPTG | + IPTG | - IPTG | + IPTG | Conclusions |
| O+ Z+ Y+ | - | + | - | + | Wild type is inducible. |
| O+ Z+ Y+/F' O+ Z- Y+ | - | + | - | + | Z+ is dominant to Z-. |
| OC Z+ Y+ | + | + | + | + | OC is constitutive. |
| O+ Z- Y+/F' OC Z+ Y- | + | + | - | + | Operator is cis-acting. |
A model of the action of lacOC in partial diploids is shown below.
The table below shows the synthesis of β-galactosidase (lacZ) and permease (lacY) in the presence and absence of the inducer IPTG. The data presented in the table are focused on a loss-of-function mutation in the repressor (lacI-).
| β-galactosidase (lacZ) | permease (lacY) | ||||
| Genotype | - IPTG | + IPTG | - IPTG | + IPTG | Conclusions |
| I+ Z+ Y+ | - | + | - | + | Wild type is inducible. |
| I- Z+ Y+ | + | + | + | + | lacI- is constitutive. |
| I+ Z+ Y+/F' I- Z+ Y+ | - | + | - | + | lacI+ is dominant to lacI-. |
| I- Z- Y+/F' I+ Z+ Y- | - | + | - | + | lacI+ is trans-acting. |
A model of the action of lacI- in partial diploids is shown below.

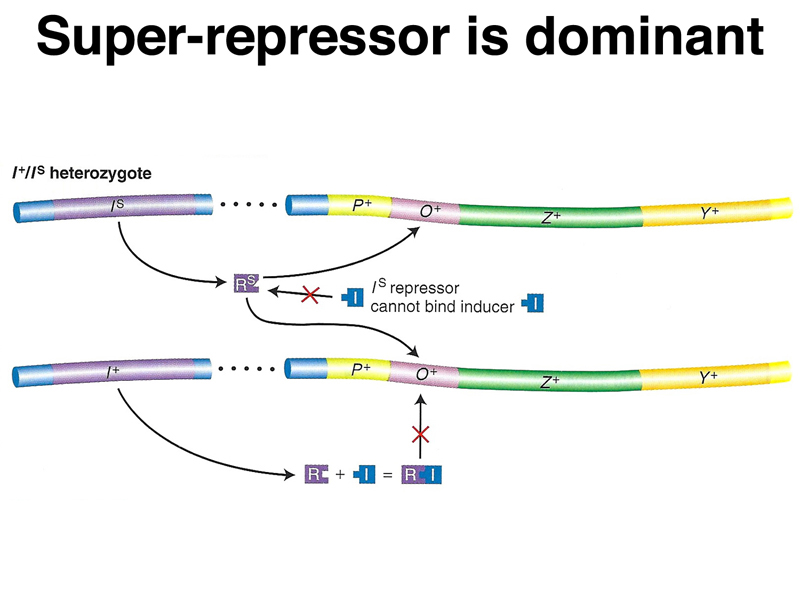
The table below shows the synthesis of β-galactosidase (lacZ) and permease (lacY) in the presence and absence of the inducer IPTG. The data presented in the table are focused on a superrepressor mutation (lacIS).
| β-galactosidase (lacZ) | permease (lacY) | ||||
| Genotype | - IPTG | + IPTG | - IPTG | + IPTG | Conclusions |
| I+ Z+ Y+ | - | + | - | + | Wild type is inducible. |
| IS Z+ Y+ | - | - | - | - | lacIS is uninducible. |
| IS Z+ Y+/F' I+ Z+ Y+ | - | - | - | - | lacIS is dominant to lacI+. |
A model of the action of lacIS in partial diploids is shown below. The repressor encoded by the superrepressor allele does not respond to the presence of inducer, either because it cannot bind inducer or because the protein does not change shape to make it unable to bind to the operator. In the presence of a normal allele and inducer, all of the superrepressor protein will be bound to all of the available operators. The superrepressor is also trans-acting.
In the last lecture, a student asked about the phenotype of cells bearing both an operator-constitutive mutation and a superrepressor allele. We know that the operator-constitutive mutation (lacOC) eliminates the ability of the repressor to bind to the operator. This would apply to the superrepressor allele (lacIS) as well, making this cell type constitutive. We can also think about what would happen in a partial diploid. The alleles in cis to the operator-constitutive mutation would be constitutive, while those in cis to the normal operator (lacO+) would be uninducible.
We leave as an exercise to the student to give the phenotype of the cells in the last two lines of the table below.
| β-galactosidase (lacZ) | permease (lacY) | ||||
| Genotype | - IPTG | + IPTG | - IPTG | + IPTG | Conclusions |
| I+ O+ Z+ Y+ | - | + | - | + | Wild type is inducible. |
| I+ OC Z+ Y+ | + | + | + | + | OC is constitutive. |
| IS O+ Z+ Y+ | - | - | - | - | lacIS is uninducible. |
| IS OC Z+ Y+ | + | + | + | + | Superrepressor can't bind the defective operator in lacOC. |
| IS O+ Z- Y+/F' I+ OC Z+ Y- | ? | ? | ? | ? | ? |
| IS OC Z- Y+/F' I+ O+ Z+ Y- | ? | ? | ? | ? | ? |
There is one additional kind of mutation in the lac operon not presented above, mutations in the promoter (lacP-). We saw last time that mutations in the promoter eliminate the binding of RNA polymerase, and therefore block transcription of all genes from that operon regardless of the genotype at lacI, lacO, lacZ, or lacY. Promoter mutations, like operator-constitutive mutations, can only be cis-acting.
Now that we know all of the kinds of mutations that there can be in the lac operon, we are ready to work some practice problems. The idea here is that you will be given the genotype of a partial diploid and asked to predict the presence or absence of lacZ and lacY proteins in the presence or absence of inducer. Often, the best way to solve these problems is to draw a cartoon, as shown below.
It would be a good idea to get good at solving these problems.
| Problem 1. Here we see a case of a promoter mutation eliminating the possibility of expression of the operon shown on top. Only the genes in the operon on the bottom have any possibility of expression. However, the bottom operon is both lacZ- and lacY-, so there won't be any expression of either gene under any circumstances and the phenotype is uninducible, as shown in the table. | |
 |
 |
| Problem 2. Here we see a case of a promoter mutation eliminating the possibility of expression of the operon shown on top. Only the genes in the operon on the bottom have any possibility of expression. The bottom operon is lacZ+ but lacY-, so there won't be any expression of lacY. There is a functional copy of lacI, so there will be normal repression. The phenotype in this case is inducible for lacZ and uninducible for lacY, as shown in the table. | |
 |
 |
| Problem 3. Here we see a case of a promoter mutation eliminating the possibility of expression of the operon shown on the bottom. Only the genes in the operon on the top have any possibility of expression. The top operon is lacZ- but lacY+, so there won't be any expression of lacZ. The operon shown on the top is lacOC, so expression of lacY will be constitutive and expression of lacZ will be uninducible, as shown in the table. | |
 |
 |
| Problem 4. The top operon is lacZ+ but lacY- with a normal operator. The bottom operon is lacZ- but lacY+ with a constitutive operator (lacOC). One of the alleles of the repressor is a superrepressor (lacIS), so expression from the top operon will be uninducible and expression from the bottom operon will be constitutive. The phenotype will be uninducible for lacZ and constitutive for lacY, as shown in the table. | |
 |
 |
Last lecture, a student asked whether nonsense or frameshift mutations in lacZ would affect the expression of lacY and lacA. Because we were trying to make the point that in a polycistronic mRNA from an operon, there are separate ribosomal binding sites and start codons for each of the genes, I answered no, that mutations in lacZ do not affect the expression of the downstream genes lacY and lacA. This is a useful answer for the purpose of understanding transcription and translation of operons, but it is not strictly correct. It is true that missense alleles of lacZ have no effect on the expression of lacY and lacA, but nonsense and frameshift alleles can have an effect on downstream genes in an operon. In fact, this was what led Jacob and Monod to consider that all three genes of the lac operon were expressed as a single transcript.
The figures and explanations below explore nonsense-mediated decay, as seen in the lac operon.
 |
This is a detailed view of regulation and gene expression in the lac operon, which we have seen before. We are going to focus on translation of the polycistronic mRNA. |
 |
Here is the mRNA from the lac operon in the process of being translated. Each gene has its own ribosome binding site and AUG codon to initiate translation. |
 |
Imagine that the transcript is now from a strain that bears a nonsense allele near the 5' end of the lacZ gene. In this case, a short peptide will be synthesized before the termination codon is reached and translation terminates. |
 |
There is now a large "bare patch" of the mRNA that is free of ribosomes. Apparently, this just doesn't look right to the cell. Recall that there is no proofreading during transcription, so the occasional transcript will have an error. One possible error is the replacement of an amino acid codon with a termination codon. It would make a lot of sense to have a cellular mechanism to eliminate these mRNAs, rather than wasting the energy involved in translating a worthless protein fragment. |
 |
The mRNA with a large patch that is free of ribosomes is degraded. This is the first example that we have seen of a regulatory mechanism that changes the half-life of mRNA. Nonsense-mediated decay (the reduction of mRNA half-life in transcripts that bear nonsense mutations) is seen in eukaryotes as well, all the way to humans. |
The trp operon of E. coli offers a useful comparison to the lac operon. The structure of the operon is shown below.

As shown above, the trp operon encodes five enzymes in the biosynthetic pathway leading to tryptophan. There is a promoter and operator as in the lac operon. In addition, there is a short leader sequence immediately following the promoter and operator. We will return to the consideration of the leader after examining the trp repressor.
As shown below, the trp repressor (actually an aporepressor, meaning that it has a necessary cofactor, tryptophan) does not bind to the operator when there is a low concentration of tryptophan. This allows transcription of the operon, increasing the cell's ability to synthesize tryptophan.

As shown below, in the presence of high concentrations of tryptophan, the tryptophan-aporepressor complex binds to the operator, repressing transcription.

In our introduction to the regulation of gene expression, we distinguished between positive and negative control, as shown in the figure below. Remember that in positive regulation, the binding of a regulatory protein to the regulatory region of a gene or operon increases transcription. In negative regulation, the binding of a regulatory protein to the regulatory region of a gene or operon decreases transcription.

We saw both kinds of regulation in the lac operon. The CAP protein and its cofactor cAMP is an activator (a positive regulator) of lac operon expression. The lac repressor is a negative regulator of lac operon expression. In the presence of lactose, the lac repressor undergoes an allosteric change in shape that makes it unable to bind to the operator, allowing transcription.

The lac repressor and the trp repressor are both negative regulators. While the lac repressor no longer binds the operator when bound to lactose, the trp repressor does not bind the operator unless it is bound to tryptophan. This difference makes sense, because the genes of the lac operon function in catabolism (burning lactose), while the genes of the trp operon function in biosynthesis (building tryptophan). It only makes sense to transcribe the lac operon in the presence of lactose, while it only makes sense to transcribe the trp operon when the cell is getting low on tryptophan.

We return to the consideration of the leader sequence of the trp operon, shown below. This is a segment following the promoter and operator, just before the beginning of the trpE gene, the most 5' gene of the operon. The leader sequence is required for attenuation, a form of transcriptional control in which RNA polymerase will either terminate transcription at the attenuator or read through to transcribe all of the genes of the trp operon.

The transcription termination signal at the attenuator is shown below. It resembles the intrinsic transcription termination signal of E. coli discussed in a prior lecture. If the 3:4 step-loop structure forms during transcription, transcription will terminate at the U shown in the figure. There is a very interesting mechanism used to control the formation of this transcription termination signal in response to the level of tryptophan.

As shown below, the sequence upstream of the transcription termination signal encodes a short peptide. There is a ribosome binding site upstream of the AUG that allows this short peptide to be translated. As indicated by the arrows, the short peptide contains two tryptophans. This is unusual, as tryptophan accounts for only about 1% of the amino acid residues in the collection of all E. coli proteins.

As shown below, downstream of the stop codon for the short peptide is a spacer region containing the sequences that encode the 3:4 step-loop structure as well as the ribosome-binding site for the trpE gene.

How does the level of tryptophan determine whether transcription will terminate at the attenuator? Recall that translation in E. coli can occur before the completion of transcription. As soon as a ribosome binding site and an AUG start codon are available on a nascent transcript, a ribosome can bind to initiate translation. The structure of the transcript from the leader region of the trp operon is shown below, with the three drawings indicating different states of the transcript.

In the drawing on the left, there has been no initiation of translation. The transcript folds into two stem-loop structures by base pairing. One of these is the 3:4 step-loop structure that acts as a transcription terminator. Transcription terminates in the run of U's as shown before.
In the drawing in the middle, translation has initiated in the presence of abundant tryptophan-charged tRNA. The ribosome proceeds through the synthesis of the leader peptide as the 3:4 step-loop structure forms to terminate transcription.
In the drawing on the right, translation has initiated at a low level of tryptophan-charged tRNA. When the ribosome reaches the two tryptophan codons, it stalls or pauses as it awaits a tryptophan-charged tRNA. The presence of a stalled ribosome in this region blocks the formation of the 1:2 stem-loop structure, because the ribosome is stalled over sequence 1. This causes an alternative stem-loop structure to form, the 2:3 stem-loop. Formation of the 2:3 stem-loop precludes the formation of the 3:4 stem-loop that acts as a transcription terminator, so transcription continues through the rest of the operon.
In this way, transcription of the trp operon is regulated at two levels: at low concentrations of tryptophan, the repressor fails to bind to the trp operator and the trp operon is derepressed. Attenuation allows an additional level of control that is sensitive to the level of charged tryptophan tRNA. The combination of these two levels of control allows the transcription of the trp operon to be regulated over a 700-fold range.
Here are some additional figures showing the mechanism of attenuation. The figure below shows the trp operon derepressed without cotranslation. Transcription is terminated at the attenuator.

In the figure below, there is a high level of charged tryptophan tRNA, and the ribosome completes the synthesis of the leader peptide without stalling. The 3:4 stem-loop forms, and transcription is terminated at the attenuator.

In the figure below, there is a low level of charged tryptophan tRNA, and the ribosome stalls during synthesis of the leader peptide. This allows the 2:3 stem-loop structure to form, blocking the formation of the 3:4 stem-loop structure that is the transcription terminator. Transcription continues through the rest of the operon.

Other biosynthetic operons in E. coli also use attenuation as a regulatory mechanism. The figure below shows the sequences of the leader peptides for the his and phe operons. Stalling of the ribosome during translation of these leader peptides blocks 1:2 stem-loop formation, causing 2:3 stem-loop formation, which prevents the formation of the 3:4 stem-loop transcription terminator.

In B. subtilis, there is no trp repressor. The trp operon of B. subtilis is regulated entirely by attenuation. In this case, the mechanism of attenuation is different. Attenuation is not dependent on cotranslation and the stalling of the ribosome during the synthesis of a leader peptide. Instead, there is a small protein called TRAP that undergoes a conformational change when bound to free tryptophan, shown below. TRAP protein that is bound to tryptophan forms an 11-mer that binds the nascent mRNA from the trp operon in the leader sequence. The TRAP 11-mer binds sequences 1 and 2, ensuring the formation of the 3:4 stem-loop structure that terminates transcription. At low concentrations of tryptophan, there is not enough TRAP 11-mer to bing the leader sequence in the mRNA, and 2:3 stem-loop formation blocks 3:4 stem-loop formation, allowing transcription of the operon.

Earlier we discussed possible levels of regulation of gene expression, and saw that the regulation of transcription was the most economical in terms of resource utiization. Attenuation is an example of the control of transcription that is almost as cheap as controlling the initiation of transcription. Using attenuation as a form of transcriptional control in addition to controlling the initiation of transcription allows the regulation of transcription over a wider range.