
| Home | Syllabus | Schedule | Lecture Notes | Extras | Glossary |

| Home | Syllabus | Schedule | Lecture Notes | Extras | Glossary |
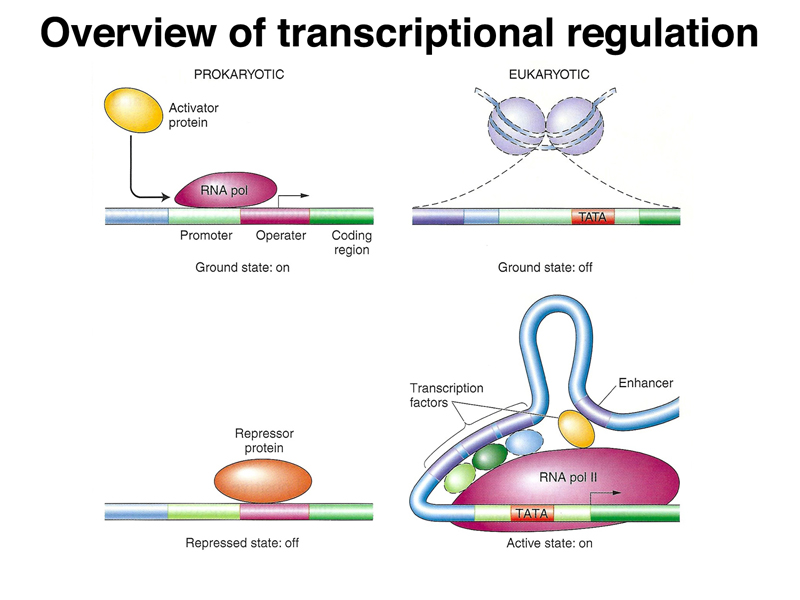
We began by once again comparing prokaryotic and eukaryotic transcriptional regulation. As shown below, in prokaryotes, the genome is essentially naked DNA with no barrier to transcription. While there are some activator proteins, like the CAP protein, that can increase the basal level of transcription under some circumstances, the default state of the prokaryotic genome is on. Repressor proteins, like the lac and trp repressor, are used to prevent transcription except under the right metabolic conditions (lactose but no glucose in the case of the lac operon, and low tryptophan levels in the case of the trp operon). In eukaryotes, the genome is packed into nucleosomes, so the ground state of transcription is off. In order for genes to be expressed, DNA must be partially unpacked from nucleosomes. Most transcriptional regulation in eukaryotes is positive: transcriptional activators that are part of a very large and complicated transcription initiation complex are the key elements of eukaryotic transcriptional regulation.
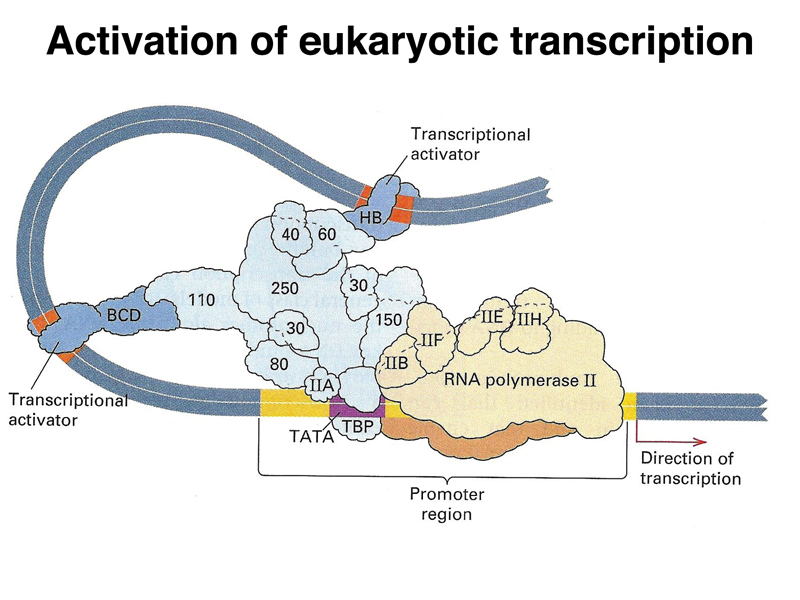
The image below is a cartoon of a eukaryotic transcription activation complex. There are three types of protein components. There is a large collection of proteins that make up a generalized transcription activator (light blue). This complex recruits RNA polymerase (beige) and its cofactors to the promoter region. There are also specific transcription factors (dark blue) that serve as activators. These interact with both the transcription activator complex and specific DNA sequences near the gene, called enhancers. By regulating the expression of these specific transcription factors, an organism can regulate the expression of a large number of genes that are under the control of a particular transcription factor. It is common for a gene to have multiple transcription factors and their enhancers that act in combination, allowing a fine-tuning of the pattern of expression of a gene or set of genes.
Eukaryotic genomes are packed into chromatin to allow large genomes to fit into nuclei of reasonable size. In addition, chromatin is packed into condensed chromosomes at a very high packing ratio in order to allow the precise partitioning of the replicated genome during cell division. There are several levels of packing used to achieve a very high packing ratio. The first three levels of packing are shown in the figure below.

The first level, naked DNA, has a "packing ratio" of 1 (the packing ratio is the ratio of the length of a particular segment of condensed chromosome relative to naked DNA). The next level is the organization of DNA into nucleosomes. Each nucleosome consists of a core of eight histone molecules (two each of H2a, H2b, H3, and H4) with DNA wound around the outside. Chromatin at this level of packing is an 11 nm fiber with a packing ratio of 6-7. The next level of packing is a 30 nm fiber with the nucleosomes packed against each other to form a tight solenoid. The packing ratio here is around 40.
The figure below summarizes the first three levels of packing, and adds several additional levels of packing that are not as well characterized. There are 70 nm loops that orignate from a scaffold; the scaffold can be further wound to make a structure just below a condensed chromosome in its packing ratio. A condensed chromosome has a packing ratio of around 10,000. Consider that naked DNA that makes up the diploid human genome has a length of about two meters, and you will see that the packing ratio of a mitotic chromosome is truly impressive.

An electron micrograph of the 11 nm fiber showing individual nucleosomes is shown below.

One level of regulation of transcription in eukaryotes is to affect the state of packing of nucleosomes. The 30 nm fiber is not accessible for transcription, while the 11 nm fiber is accessible. The transition from one state to the other is affected by posttranslational modification of histone "tails" that hang off the main part of each histone. This is shown in the cartoon below.

Sites of modification of the amino-terminal tails of the core histones are shown below. The modifications include acetylation of many different lysine residues, methylation of arginine and lysine residues, and phosphorylation. Specific modifications are made or removed by enzymes specialized for a particular modification.

What is the effect of these modifications? Histones stick to DNA because they are positively charged, allowing them to be attracted to the negatively-charged sugar-phosphate backbone of DNA. Acetylation of histone tails makes them less positively charged, and tends to promote a looser configuration of chromatin compared to the state in which the acetylation is not present.
A model of the 30 nm fiber is shown below. The 30 nm solenoid requires the association of histone H1 with the octomeric histone core as shown.

One method of exploring chromatin structure experimentally is nuclease digestion, as shown below. The 30 nm fiber is relatively resistant to DNaseI, while the 11 nm fiber is 100x more sensitive. An entire genome's worth of chromatin can be treated with DNaseI at once, then regions of interest can be examined using molecular techniques that we have not yet discussed. In this way, it is possible to examine the regulatory region of an individual gene to survey the state of chromatin condensation.

This allows us to characterize the regulatory region of a specific gene, looking for DNaseI-hypersensitive sites that contain regulatory sequences. A cartoon of such findings is shown below. As we would expect, the promoter region of the gene is DNaseI-hypersensitive. Enhancer regions, which can be located 100 kb away, are also DNaseI-hypersensitive. Enhancers do not need to be located upstream of a gene. They can be located downstream, or even in one of the introns.

We will take up the remodeling of chromatin to expose regulatory sites in the next lecture. For now, we will focus on the regulation of the state of large stretches of chromatin. As mentioned before, this is influenced by the state of acetylation of histone tails, as shown below.

Histone acetyltransferases (HATs) add acetyl groups to histone tails, promoting an open chromatin configuration (the 11 nm fiber). Histone deacetylases (HDAC) remove acetyl groups from histone tails, promoting an inaccessible chromatin configuration (the 30 nm fiber).
Examination of nuclei from many different eukaryotes reveals that the nucleus contains two types of chromatin: euchromatin, which is relatively loosely condensed and transcriptionally active, and heterochromatin, which is more tightly condensed and transcriptionally inactive. Heterochromatin also tends to replicate later in S phase than does euchromatin.
In condensed chromosomes, we typically see heterochromatin around the centromere. Often about a third of a condensed chromosome is pericentric heterochromatin. There is also usually telomeric heterochromatin, as shown in the figure below.

The figure below points out some additional differences between euchromatin and heterochromatin: heterochromatin contains satellite sequences, short sequences repeated millions of times, as well as middle-repetitive transposible genetic elements. It has a very low density of transcribed genes. The figure also points out specific histone modifications that differ between euchromatin and heterochromatin: methylation of lysine 9 of histone H3 in heterochromatin, and methylation of lysine 4 of histone H3 in euchromatin.

We understand quite a bit about the formation of heterochromatin in Drosophila as a result of genetic analysis. In order to introduce the subject, we present the life cycle of Drosophila in the figure below.

Adult Drosophila lay eggs, which hatch into larvae. The larvae feed and grow, undergoing three molts before the third-instar larva enters pupation. During pupation, about 90% of the cells in the larva are destroyed. Much of the adult body is derived from groups of cells set aside in the larva. Because many of the larval cells are destined to be destroyed and won't need to divide again, they are terminally differentiated. In particular there is a set of "salivary glands" made up of cells that have undergone endomitotic replication (replication without cell division) to amplify the genome. These cells make a glue protein that the larva uses to attach itself to a substrate for pupation.
The amplification of the genome in the salivary glands (called polytenization) is uneven. Euchromatin amplifies, but most of the heterochromatin hardly amplifies at all. As shown in the figure below, this results in a chromosome set in which the five major chromosome arms (X, 2L, 2R, 3L, and 3R) are seen emerging from the pooled heterochromatin, called the chromocenter. The euchromatin of each chromosome arm is amplified to about 1,000 copies. The mitotic chromosomes (not to scale) are shown as an inserted figure in the upper left.

The lack of amplification of heterochromatin in polytene tissue is another distinguishing characteristic of heterochromatin in Drosophila.
A large number of chromosome rearrangements have been recovered in Drosophila. Some chromosome rearrangements that have a one breakpoint in euchromatin and one in heterochromatin exhibit position-effect variegation, as shown in the images below. There is an X chromosome inversion that has one breakpoint near the site of the white gene and the other breakpoint in the pericentric heterochromatin of the X chromosome. Flies bearing this inversion show a mottled or variegated expression of the white gene, as shown in the photographs and drawings.


Position-effect variegation in this case is interpreted to be a spreading of heterochromatin formation into the region including the white gene. In the uninverted chromosome, there is a barrier of some kind that marks the euchromatin-heterochromatin boundary. When a new euchromatin-heterochromatin junction is created as a consequence of the inversion, it lacks a boundary, and some of the euchromatin, including that containing the white gene, becomes heterochromatic and transcriptionally inactivated. This phenomenon provides an opportunity to investigate the formation of heterochromatin.
Position-effect variegation is sensitive to genetic and environmental modifiers. For example, as shown below, X0 males bearing the variegating X chromosome show more inactivation of white, while XYY males bearing the variegating X chromosome show less.

The Y chromosome is almost entirely heterochromatic, so one way to look at these results is that adding an extra Y chromosome "soaks up" the cell's capacity to make heterochromatin, and position-effect variegation is suppressed. Similarly, removing a Y chromosome to make an X0 male gives the cell added capacity to make heterochromatin, and position-effect variegation is enhanced.
There is nothing special about the Y chromosome in this regard. Adding or removing autosomal heteochromatin using deletions and duplications has a similar effect.
It is possible to identify mutations that suppress or enhance position-effect variegation, as shown below. These mutations are typically dominant. There are over 100 genes the act as suppressors or enhancers of position-effect variegation.

As shown below, suppressors of position-effect variegation are additive. In this case, the generic Su(var) name has been replaced by names that describe the molecular function of the mutated gene. The two suppressors are hdac1 and hdac3, both histone deacetylases. It makes sense that reducing the activity of a histone deacetylase or two would reduce the cell's capacity to make heterochromatin, because acetylated histones are associated with decondensed chromatin.

Another suppressor of position-effect variegation is Su(var)205, also known as HP1 or heterochromatin protein 1. As shown below, antibodies against the HP1 protein stain the chromocenter of polytene chromosomes. In these images, the site of antibody binding is detected by a secondary antibody that has been chemically modified to carry a fluorescent molecule.

The figure above shows that antibodies to HP1 protein stain the chromocenter, the heterochromatic fourth chromosome, and portions of the other chromosome arms.

The figure above shows that HP1 and a different protein, HP2, are both associated with heterochromatin. Mutations that reduce the expression of these proteins suppress position-effect variegation, suggesting that HP1 and HP2 aare required for the formation of heterochromatin.
These findings are summarized in the model below. Acetylated histones are associated with euchromatin, so mutations that reduce the function of histone deacetylases are suppressors of position-effect variegation. Histones that are methylated at particular positions are associated with heterochromatin. The HP1 protein binds to these methylated histones and recruits a histone methylase, allowing heterochromatin to spread to nearby regions until a boundary element is encountered. In a chromosome rearrangement like the X-chromosome inversion that we have discussed, the novel euchromatin-heterochromatin junction lacks a boundary element, so heterochromatic inactivation of the white gene takes place in some cells.

The investigation of position-effect variegation has given us some insights into the formation of euchromatin and heterochromatin, but this is not very interesting until we can see that it applies to a biological problem of more general importance. Fortunately, the state of chromosome condensation has to do with dosage compensation in both mammals and Drosophila.
This leads us to compare the rules for sex determination in Drosophila and humans that we described in lecture 6.
| Drosophila female | Drosophila male | Human female | Human male |
| XX | XY | XX | XY |
| XXY | X0 (sterile) |
X0 (Turner) |
XXY (Klinefelter) |
| XXX (metafemale) |
XYY | XXX | XYY |
| In Drosophila, individuals with a single X are male. The Y chromosome is not involved in sex determination. |
In humans, individuals with at least one Y chromosome are male. The X chromosome is not involved in sex determination. |
||
We had previously discussed that in humans, the presence of one or more Y chromosomes determines maleness. In the absence of a Y chromosome, female development takes place. In humans and other mammals dosage compensation is achieved in XX females by the inactivation of one of the two X chromosomes early in development, when there are about 50 - 100 cells. After this event, the same X chromosome is inactive in all of the daughter cells derived from a particular cell. This leads to the tortoiseshell and calico cats that we have previously used as examples; these cats are heterozygous for the X-linked Orange allele. Cells where the Orange X chromosome is inactive produce black fur, and cells where the other X chromosome is inactive produce orange fur. The cells of female mammals contain a Barr body, a heterochromatic X chromosome.
Klinefelter males (XXY) also have Barr bodies, demonstrating that phenotypic sex and dosage compensation are not completely correlated in mammals.
These observations on X chromosome inactivation in mammals show that a particular chromosome might or might not be heterochromatic. Heterochromatin of this kind has been called facultative heterochromatin, while the heterochromatin around the centromere has been called constitutive heterochromatin. The phenomenon of position-effect variegation shows that any sequence can become heterochromatic.
We know that the mechanism of dosage compensation must be different in Drosophila. In the crosses using the sex-linked white gene, we saw that w is recessive: females heterozygous for white (w/+) are phenotypically identical to females homozygous for the wild-type allele (+/+). Unlike humans and cats, Drosophila does not achieve dosage compensation by inactivating one X in females.
We also see a difference in the ability of humans and Drosophila to tolerate additional X chromosomes. In humans, XXX, XXXX, and so on, are normal females with extra Barr bodies in each cell. In Drosophila, XXX individuals are poorly viable metafemales, and XXXX are never viable.
The key insight into the Drosophila dosage compensation system came from the recovery of male-specific lethals. These are recessive mutations that are lethal to males but not to females. There are five genes known: mle, msl-1, msl-2, msl-3, and mof. All of these genes have been analyzed at the molecular level. The genes and their known functions are summarized below.
| Gene | Protein function |
| mle | dsRNA binding |
| msl-1 | chromatin binding |
| msl-2 | chromatin binding (zinc finger) |
| msl-3 | methylated histone binding |
| mof | histone acetyltransferase |
The protein products of these genes are known to form a complex that is associated with the X chromosome in males. The figure below shows polytene chromosomes from a male larva stained with a DNA stain (left) or with fluorescent antibodies to MSL-3 (right).

The figure above shows that the MSL-3 protein specifically associates with the X chromosome in males. Similar results are found for the protein products of the other genes. In male larvae that are dying because they are homozygous for a loss-of-function allele of one of these genes, the other proteins are not associated with the X chromosome, suggesting that all of the proteins form a complex that binds to the X chromosome of males. The presence of a histone acetyltransferase (MOF) in this complex suggests that the mechanism of dosage compensation in Drosophila is hypertranscription of the X chromosome due to an altered chromatin conformation in males.
The figure below shows direct evidence of this. The panels on the left show staining of DNA (blue) or antibodies to MSL1, MSL2, MLE, and acetylated histone H4 (yellow or orange) in polytene chromosomes from normal male larvae. The panels on the right show show staining of DNA (blue) or antibodies to MSL1, MSL2, MLE, and acetylated histone H4 (yellow or orange) in polytene chromosomes from male larvae that are dying because they are hemizygous for a loss-of-function allele of mof.

You can see that males dying from the mof mutation have reduced association of MSL1 and MSL2 protein with the X chromosome, and that there is no MLE on the X chromosome of mutant males. Acetylated histone H4 is also absent from the X chromosome of mutant males.
The chromatin configuration of the male X chromosome in Drosophila is novel. It is euchromatic, but looser than autosomal euchromatin.
How many chromatin states are there? The modENCODE project has systematically investigated the configuration of chromatin along the entire Drosophila genome in great detail. This is done using chromatin immunoprecipitation, an experimental technique that uses an number of distinct steps, described below.
The results from a large number of experiments are summarized in the image below. Statistical analysis suggests nine distinct chromatin states.

Here are the results mapped to the Drosophila genome. Notice that heterochromatin is a distinct state that can be described by a particular combination of histone modifications (generally reduced methylation and acetylation, but enrichment for methylation of histone H3). The male X chromosome is enriched for a particular chromatin state (acetylation of histone H4 at lysine 16).

In conclusion, regulation of chromatin states is an important part of the regulation of gene expression in Drosophila, humans, and other eukaryotes.