| Student name: | Candice Cooper | |
| Student email: | cfcooper@unm.edu | |
| Faculty advisor: | Dr. Paul Szauter | |
| College/University: | University of New Mexico | |
Project Details | ||
| Project name: | dananassae_3Lcontrol_Jan2013_fosmid_2392K12 | |
| Project species: | Drosophila ananassae | |
| Date of submission: | ||
| Size of project in base pairs: | 38,496 | |
| Number of genes in project: | 2 | |
Does this report cover all genes and all isoforms or is it a partial report?
All genes and isoforms
Complete the following Gene Report Form for each gene in your project. Copy and paste the sections below to create as many copies as needed. Be sure to create enough Isoform Report Forms within your Gene Report Form for all isoforms.
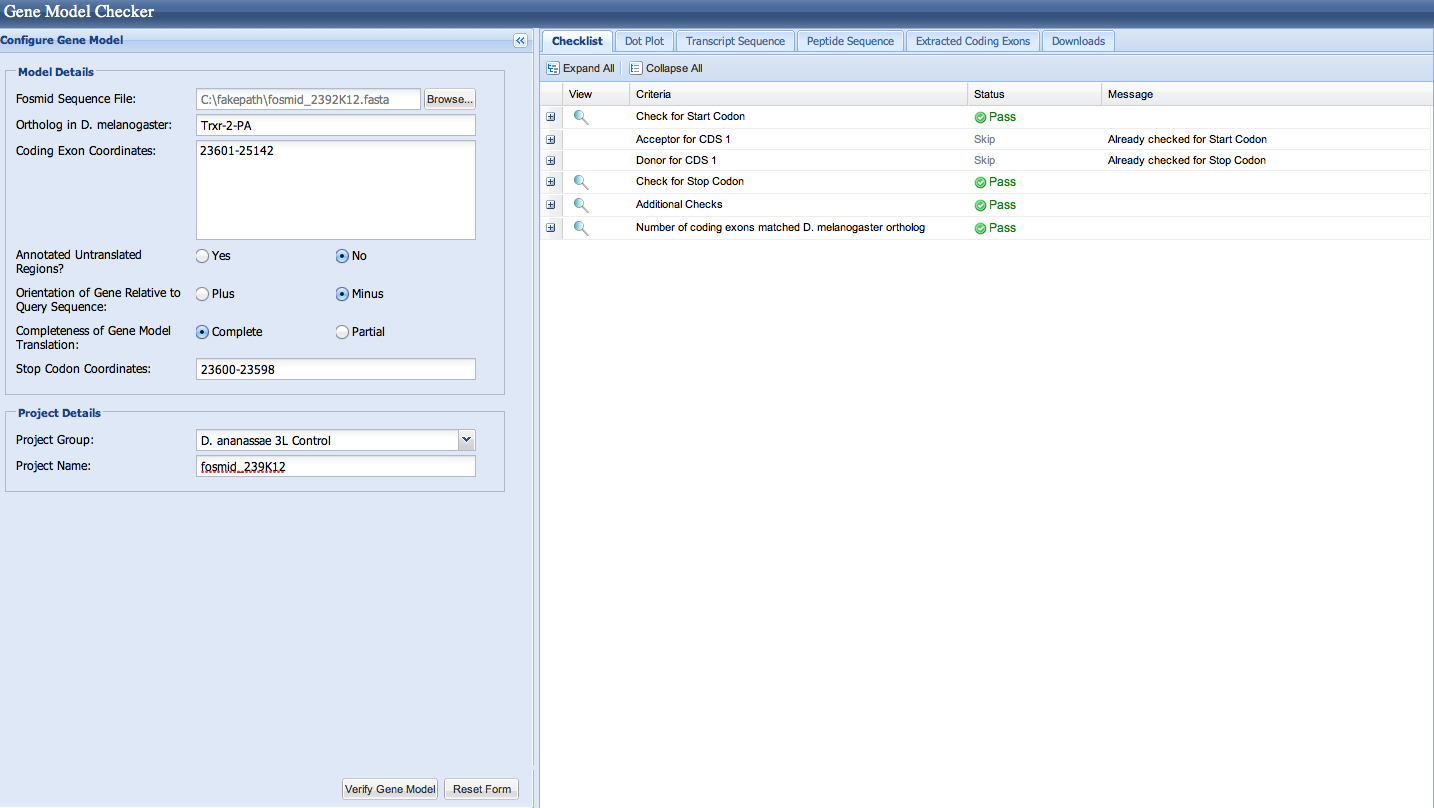
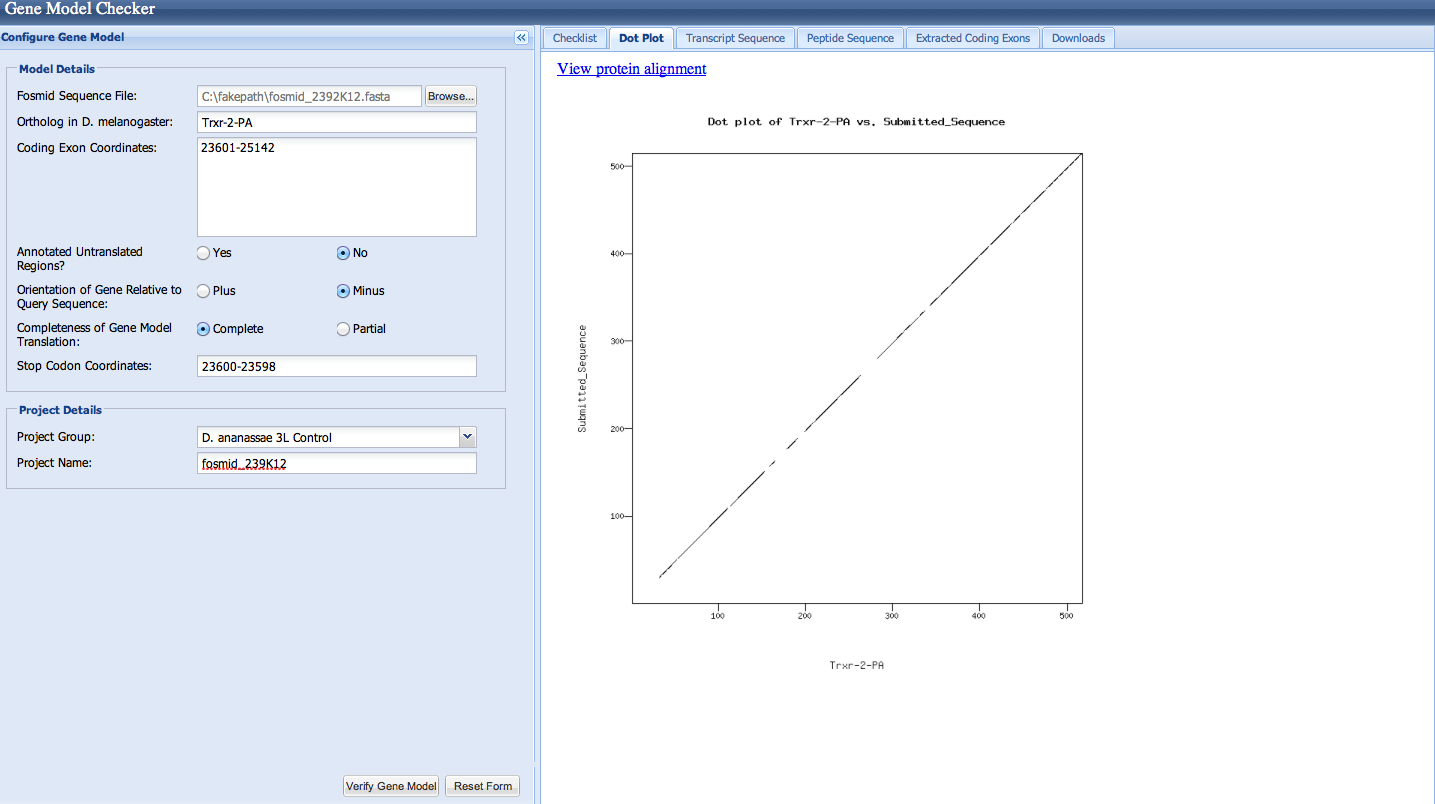
| Gene name (i.e. D. mojavensis eyeless): | Thioredoxin reductase 2 |
| Gene symbol (i.e. dmoj_ey): | dana_Trxr-2 |
| Approximate location in project (from 5' end to 3' end): | 25,142-23,601 |
| Number of isoforms in D. melanogaster: | 1 |
| Number of isoforms in this project: | 1 |
| Name of unique isoform based on coding sequence | List of isoforms with identical coding sequences |
| Trxr-2-PA | |
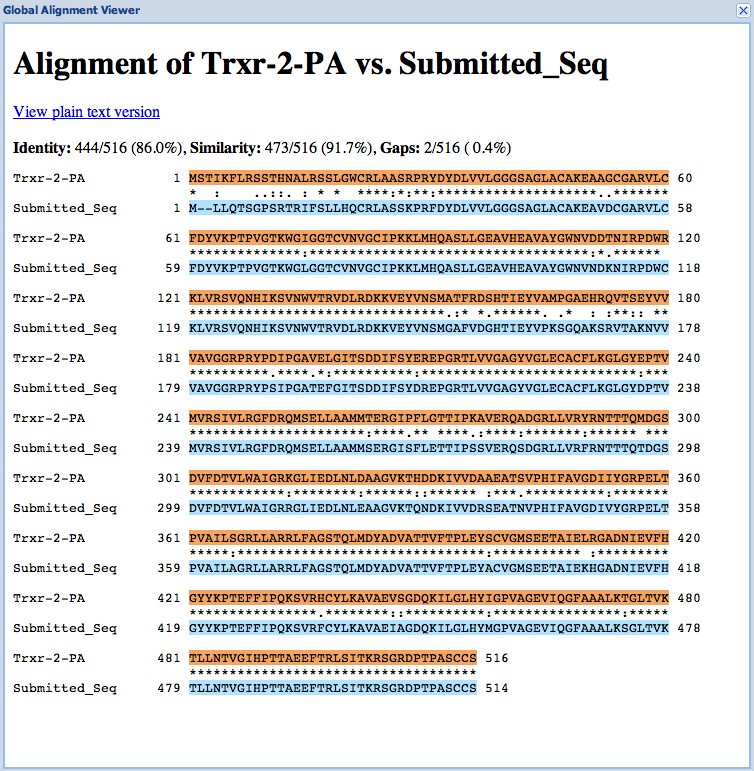
Gene-isoform name (i.e. dmoj_ey-PA): dana_Trxr-2-PA
Names of the isoforms with identical coding sequences as this isoform: none
Is the 5' end of this isoform missing from the end of project: no
If so, how many exons are missing from the 5' end: N/A
Is the 3' end of this isoform missing from the end of the project: no
If so, how many exons are missing from the 3' end: N/A

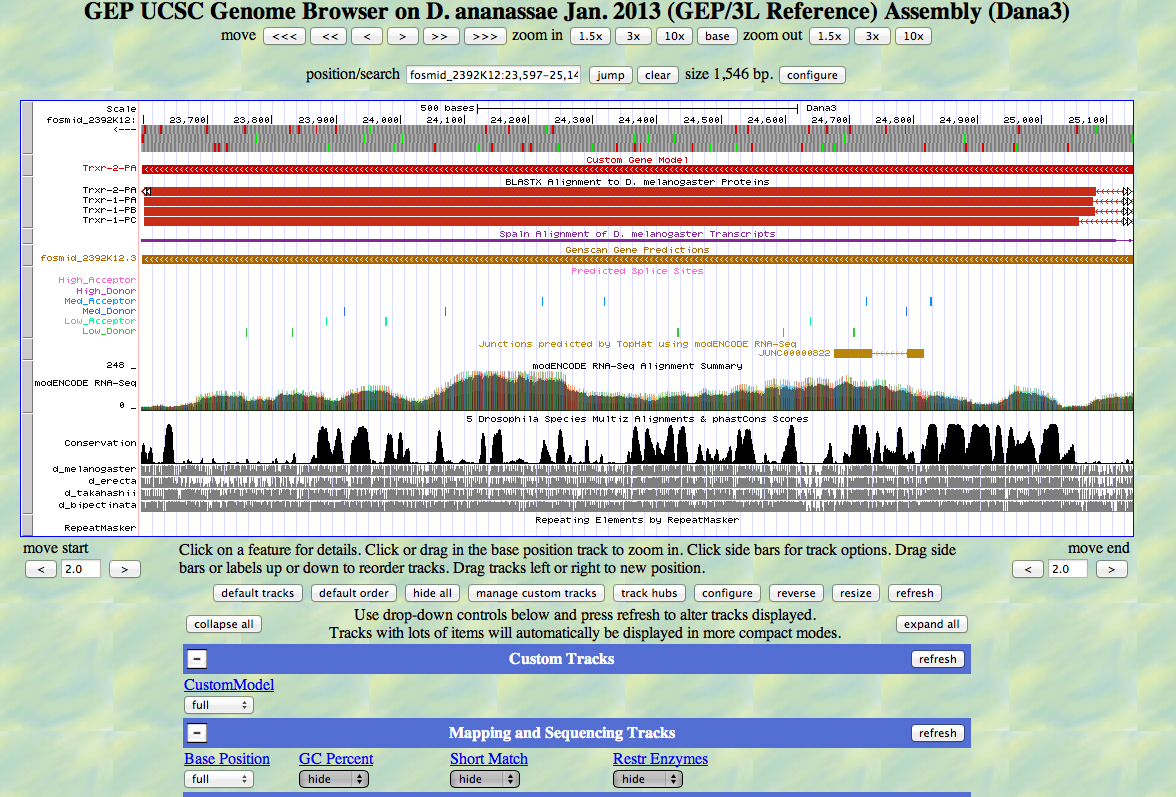
Trxr-2 has had a microinversion in D. ananassae from D. melanogaster. It is on the (-) strand in D. ananassae and the (+) strand in D. melanogaster.
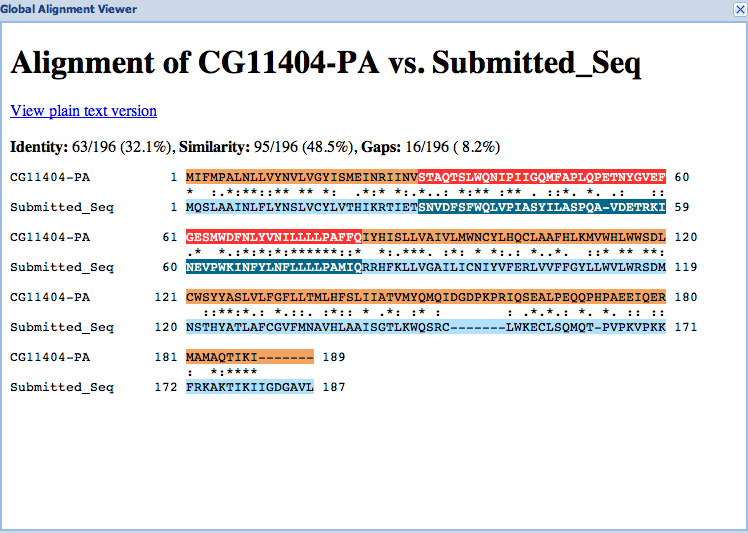
| Gene name (i.e. D. mojavensis eyeless): | CG11404 |
| Gene symbol (i.e. dmoj_ey): | dana_CG11404 |
| Approximate location in project (from 5' end to 3' end): | 21671-22338 |
| Number of isoforms in D. melanogaster: | 2 |
| Number of isoforms in this project: | 2 |
| Name of unique isoform based on coding sequence | List of isoforms with identical coding sequences |
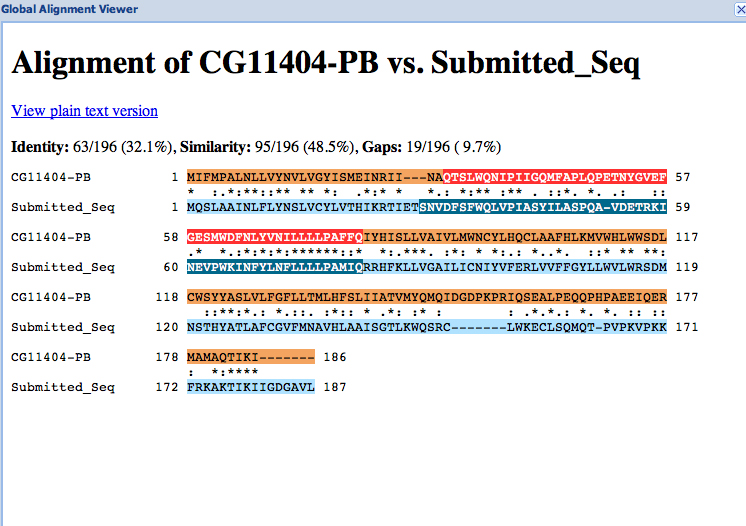
| CG11404-PA | CG11404-PB |
Gene-isoform name (i.e. dmoj_ey-PA): dana_CG11404-PA
Names of the isoforms with identical coding sequences as this isoform: CG11404-PB
Is the 5' end of this isoform missing from the end of project: no
If so, how many exons are missing from the 5' end: N/A
Is the 3' end of this isoform missing from the end of the project: no
If so, how many exons are missing from the 3' end: N/A
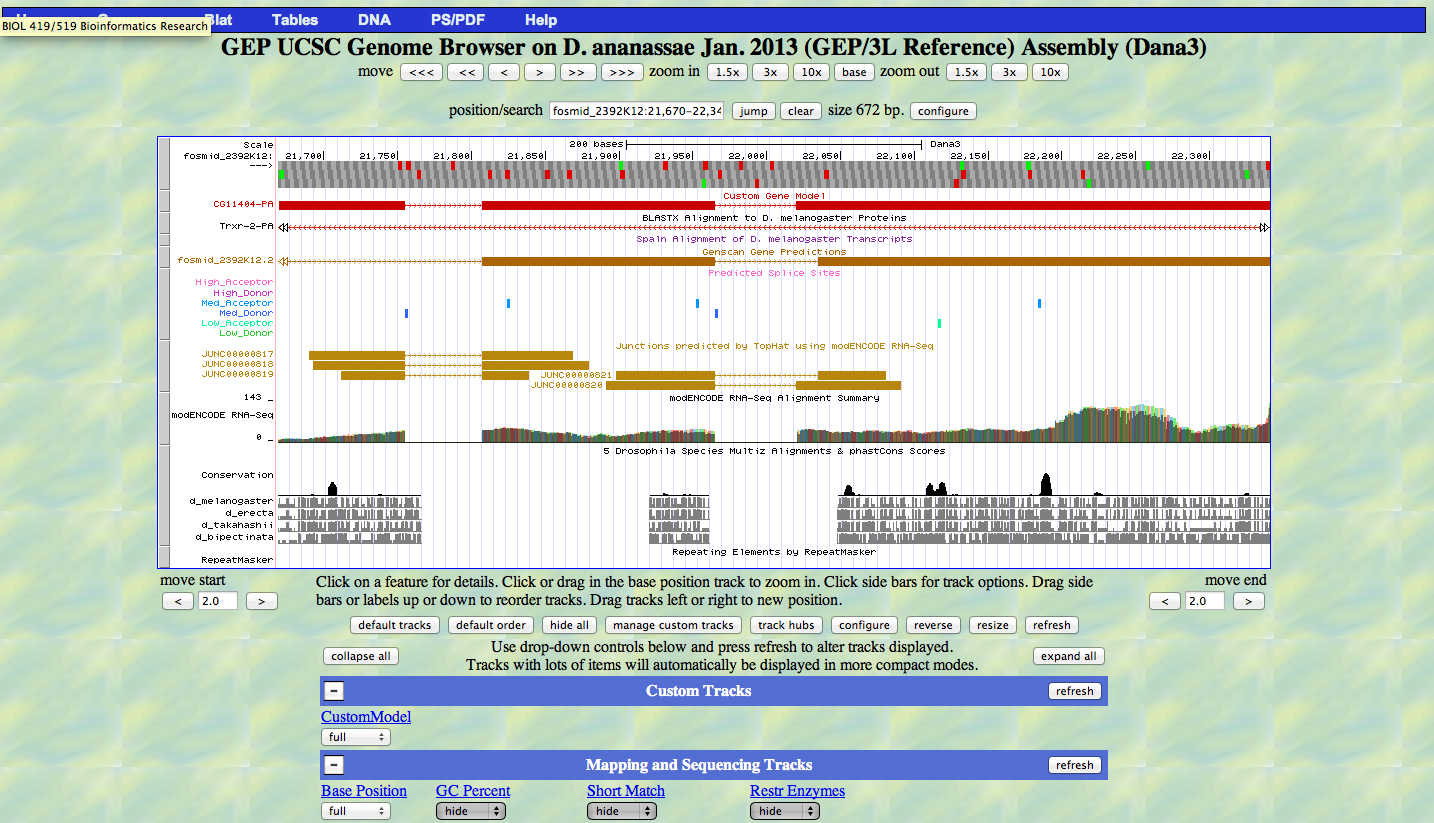
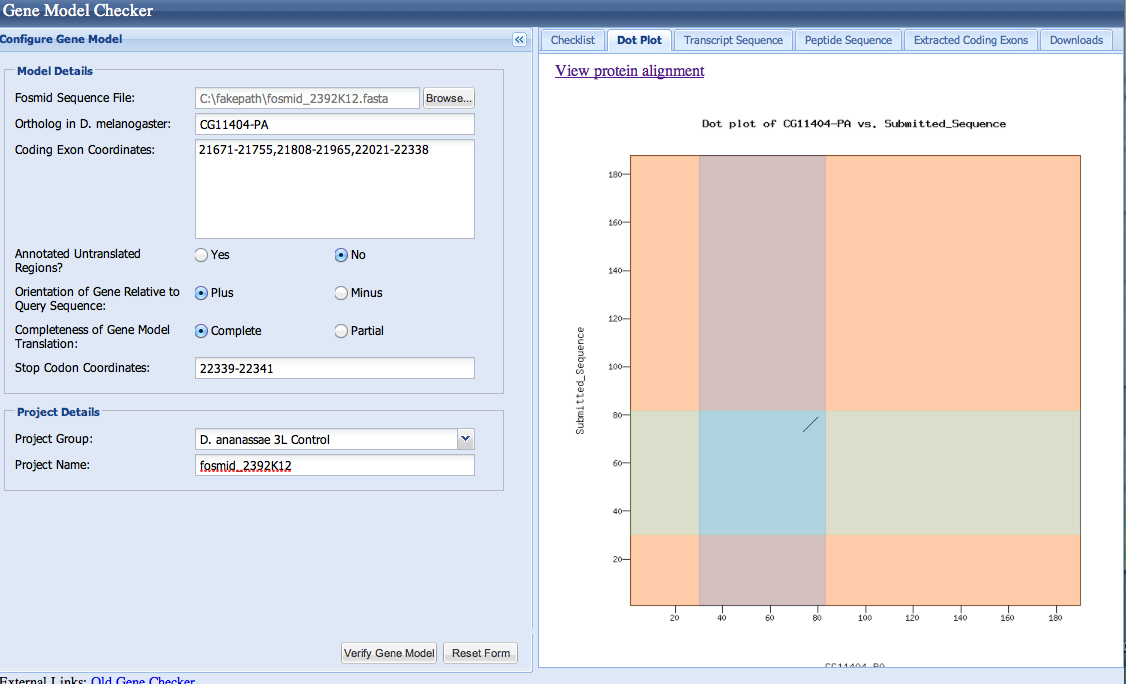
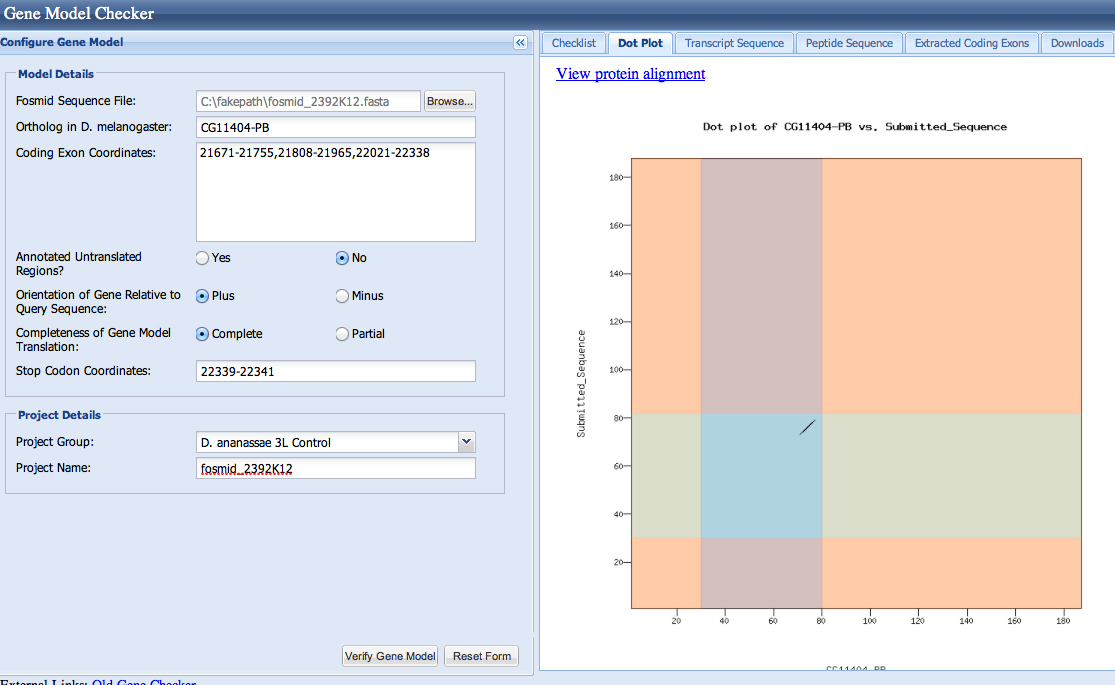
For each project, you should prepare the project GFF, transcripts and peptide sequence files (for ALL isoforms) along with this report. You can combine the individual files generated by the Gene Model Checker into a single file using the Annotation Files Merger.
The Annotation Files Merger also allows you to view all the gene models in the combined GFF file within the Genome Browser. Please refer to the Annotation Files Merger User Guide for detail instructions on how to view the combined GFF file on the Genome Browser (you can find the user guide under “Help” -> “Documentations” -> “Web Framework” on the GEP website at http://gep.wustl.edu).
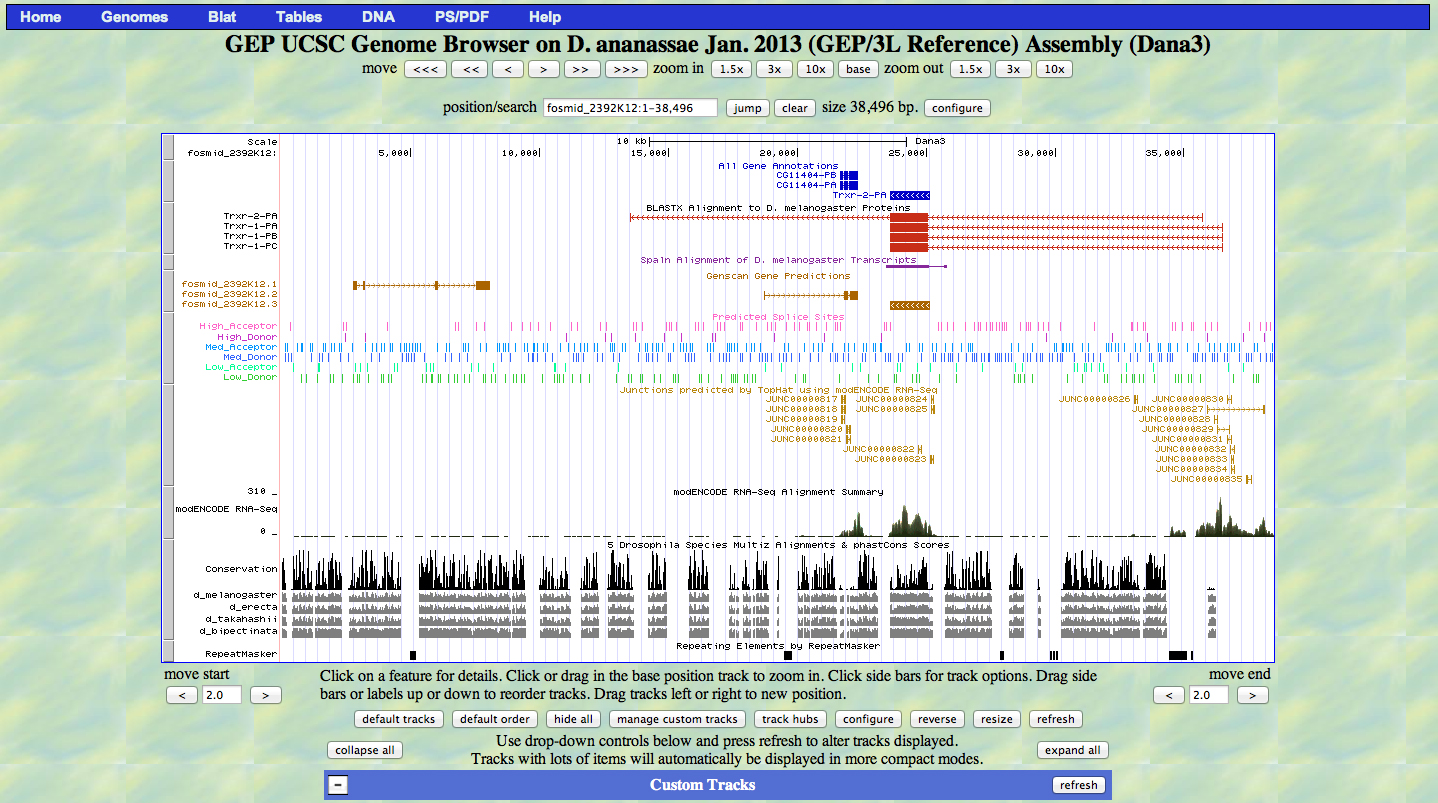
Paste a screenshot (generated by the Annotation Files Merger) with all the gene models you have annotated in this project.
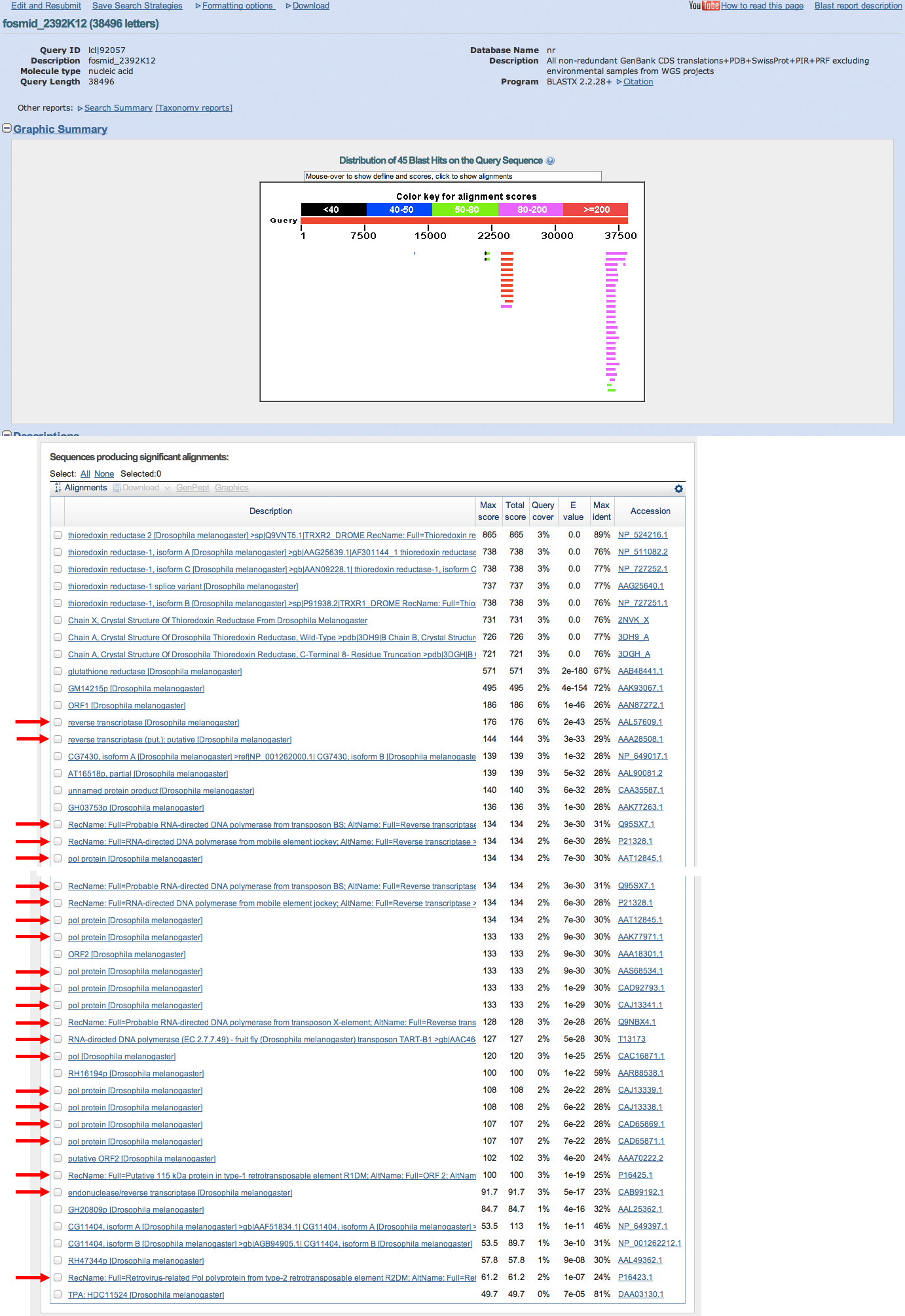
For each region of the project with gene predictions that do not overlap with putative orthologs identified in the BLASTX track, perform a BLASTP search using the predicted amino acid sequence against the non-redundant protein database (nr). Provide a screenshot of the search results. Provide an explanation for any significant (E-value < 1e-5) hits to known genes in the nr database and why you believe these hits do not correspond to real genes in your project.
3 genscan gene predictions, 1 invalid
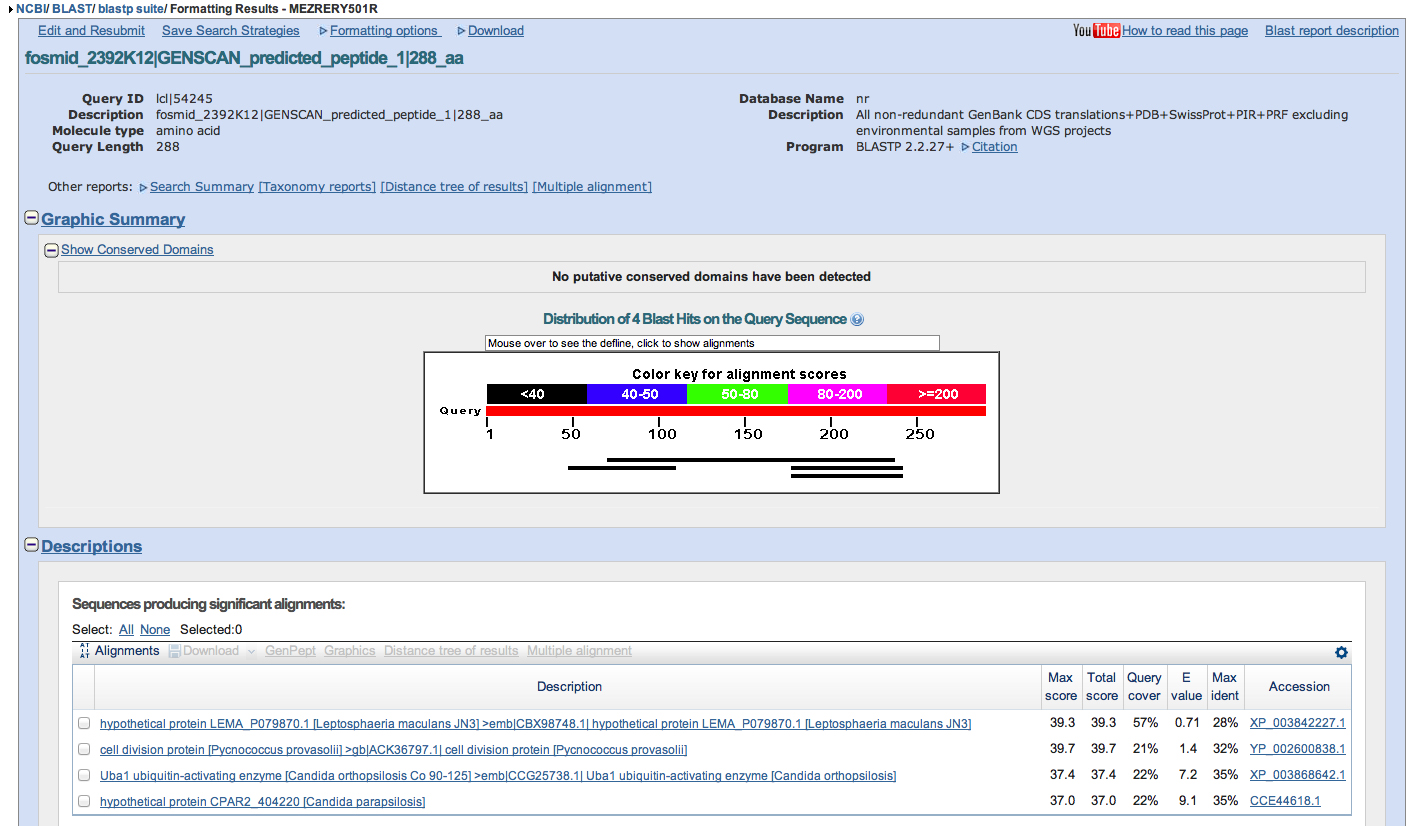
no significant hits against entire nr database, invalid gene prediction
Wolbachia repeats seen around 4967-5194, 30078-30129, 19501-19615, 34435-3511
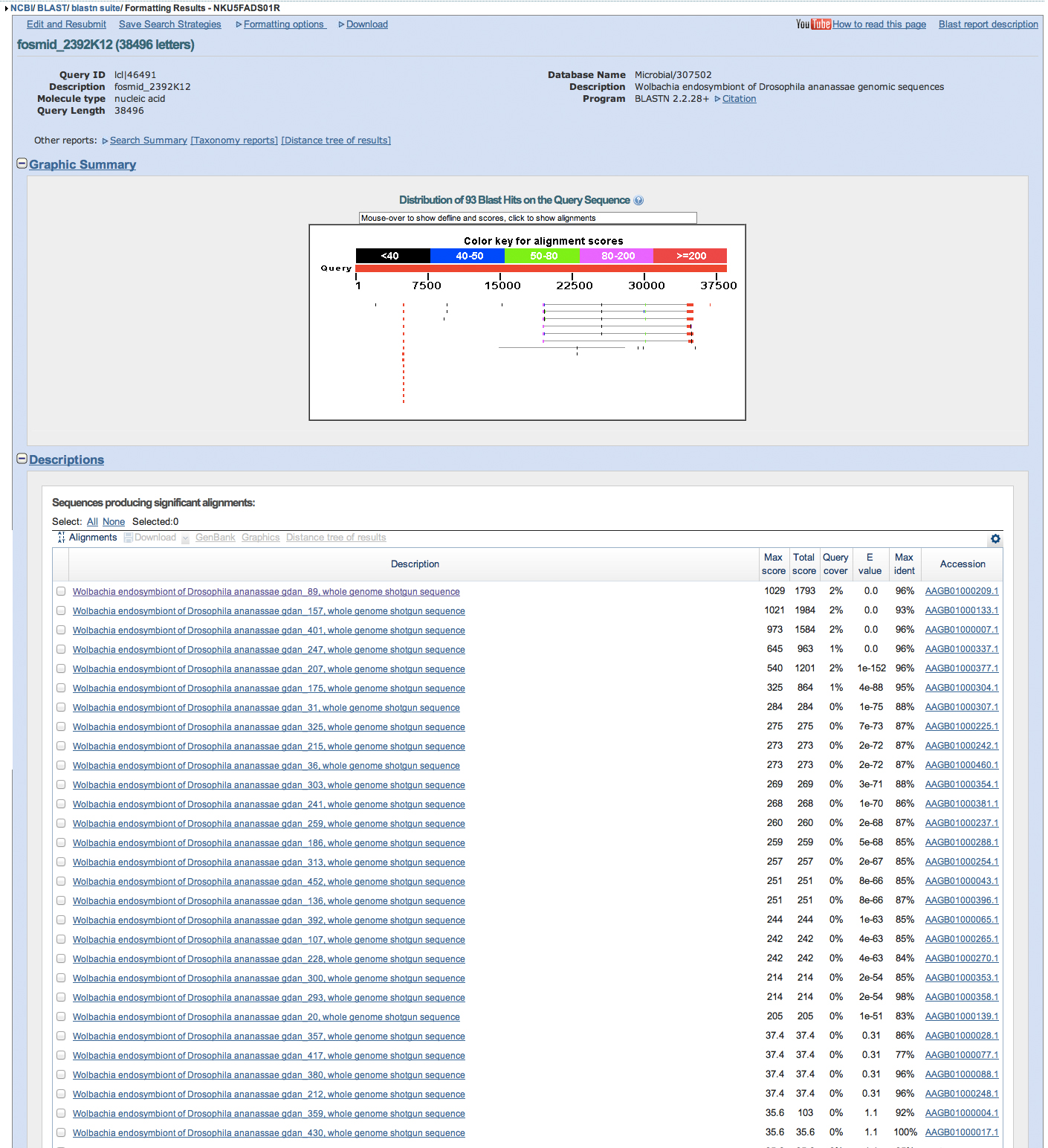
retroviral repeats seen around ***35,000***