This is an example of how to write the GEP Annotation Report. Submission of the GEP Annotation Report and the associated files is required to complete this course. The GEP Annotation Report template is posted on this website, and is followed here.
The GEP Annotation Report asks you to identify yourself as follows:
| Student name: | Eugene Hunter | |
| Student email: | genehunter@unm.edu | |
| Faculty advisor: | Dr. Paul Szauter | |
| College/University: | University of New Mexico | |
Project Details | ||
| Project name: | dananassae_3Lcontrol_Jan2013_fosmid_2748I18 | This is the name of the folder containing your fosmid sequence and other files. |
| Project species: | Drosophila ananassae | |
| Date of submission: | April 1, 2013 | |
| Size of project in base pairs: | 39,531 | You can obtain this number by viewing your fosmid in the GEP UCSC Genome Browser and noting the end coordinate. |
| Number of genes in project: | 3 | This is the final number of genes you have found. GENSCAN models that are not valid do not count as genes. We are giving the number as three, because this fosmid contains CS-2, Act79B, and CG7470. |
Does this report cover all genes and all isoforms or is it a partial report?
This example is a partial report; you should cover all of the genes on your fosmid.
If this is a partial report because different students are working on different regions of this sequence, please report the region of the project covered by this report:
from base 1 to base 8902
We have given the base count from base 1 (the left edge of the fosmid) to base 8902, the last base of the stop codon in CS-2.
Complete the following Gene Report Form for each gene in your project. Copy and paste the sections below to create as many copies as needed. Be sure to create enough Isoform Report Forms within your Gene Report Form for all isoforms.
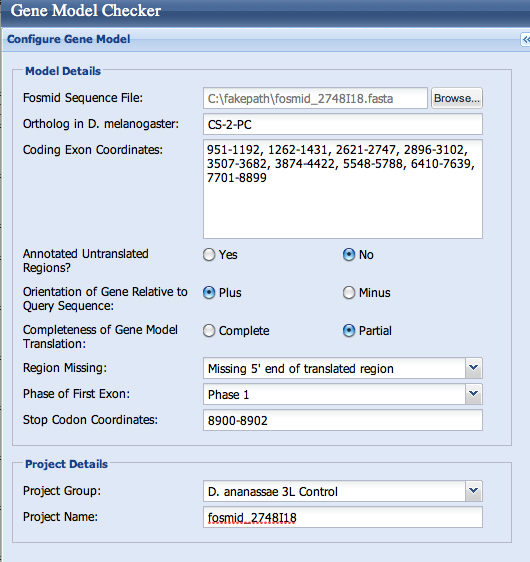
| Gene name (i.e. D. mojavensis eyeless): | D. ananassae Chitin Synthetase 2 |
| Gene symbol (i.e. dmoj_ey): | dana_CS-2 |
| Approximate location in project (from 5' end to 3' end): | 1 - 8902 |
| Number of isoforms in D. melanogaster: | 2 |
| Number of isoforms in this project: | 1 |
Complete the following table for all the isoforms in this project:
If you are annotating untranslated regions then all isoforms are unique (by definition)
| Name of unique isoform based on coding sequence | List of isoforms with identical coding sequences |
| CS-2-RC | none |
Isoform report form
Complete this report form for each unique isoform listed in the table above (copy and paste to create as many copies of this Isoform Report Form as needed):
Gene-isoform name (i.e. dmoj_ey-PA): dana_CS-2-PC
Names of the isoforms with identical coding sequences as this isoform: none
Is the 5' end of this isoform missing from the end of project: yes
If so, how many exons are missing from the 5' end: 1
Is the 3' end of this isoform missing from the end of the project: no
If so, how many exons are missing from the 3' end: none

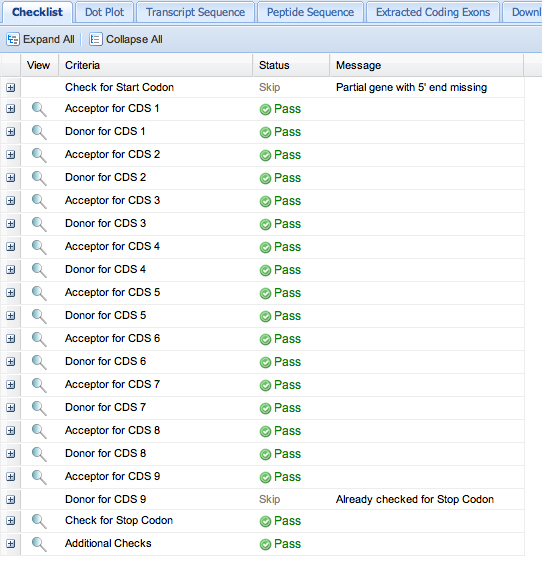
First, save the GFF file from the Gene Model Checker.
Here is the GFF file from the Gene Model Checker:
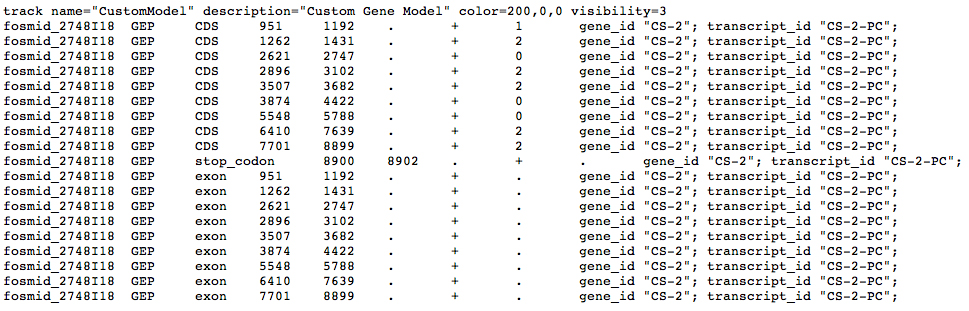
Please see the Instructions for uploading your custom track to the GEP UCSC Genome Browser.
Go to the GEP Genome Browser Gateway and specify the fosmid. Click the button indicated by the red arrow below to add custom tracks.

In the screen that appears, use the Choose File button indicated by the red arrow to upload your GFF file. Click the Submit button.
Capture a screenshot of your gene model shown on the Genome Browser for your project; zoom in so that only this isoform is in the screenshot. Include the following evidence tracks in the screenshot if they are available.
1. A sequence alignment track (D. mel Protein or Other RefSeq)
2. At least one gene prediction track (e.g. Genscan)
3. At least one RNA-Seq track (e.g. RNA-Seq Alignment Summary)
4. A comparative genomics track
(e.g. Conservation, D. mel. Net Alignment, 3-way, 5-way or 7-way multiz)
Paste the screenshot of your gene model as shown on the Genome Browser below:

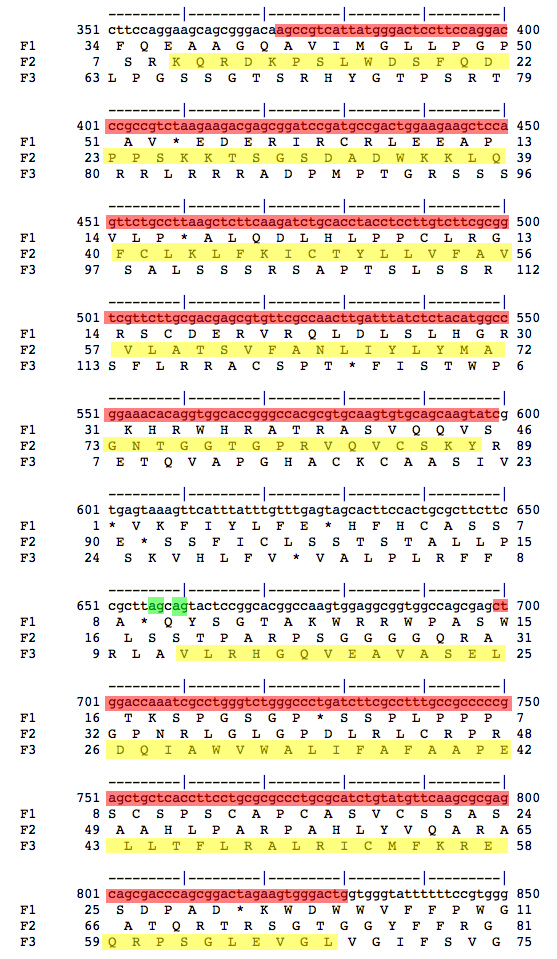
We want to decide whether Exon 3 or Exon 4 was lost in D. ananassae. We retrieve genomic sequences for D. melanogaster from FlyBase and use our alignments to select the relevant portion of the sequence of the D. ananassae fosmid.
We use the EMBOSS toolkit, selecting showorf as the relevant tool. The screenshots shown below were colorized in Photoshop to highlight the features under discussion.
Dmel exons 2 & 3/4 |
Dana exons 2 & 4 |
 |
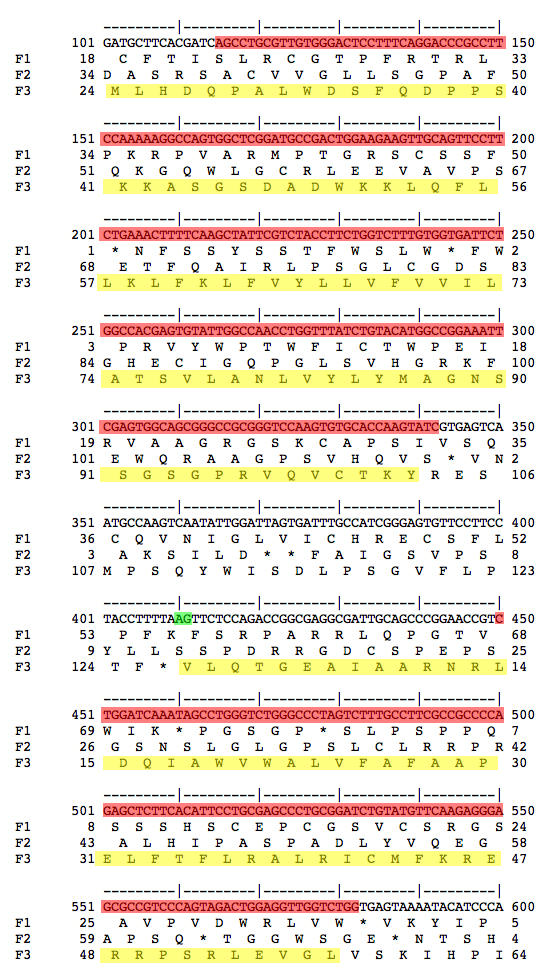 |
| Genomic sequence for Exons 2 and 3/4 from D. melanogaster and D. ananassae are shown. The amino acid sequences for the coding sequences are highlighted in yellow. The nucleotide sequences that align between D. melanogaster Exons 2 and 3/4 and genomic sequence from D. ananassae are shown highlighted in red. Splice acceptors at the 5' end of Exon 3/4 are shown highlighted in green. The most parsimonious interpretation is that D. ananassae has lost the first splice acceptor and retained the second. Therefore, D. melanogaster Exon 3 is not used in D. ananassae, only Exon 4. This means that D. ananassae lacks an isoform corresponding to CS-2-RD, and only has a single isoform corresponding to CS-2-RC in D. melanogaster. | |