In order to analyze stop codon readthrough in the Ten-m gene, we need to collect the genomic sequence of the last coding exon and some of the 3' sequence from Ten-m and control genes.
| D. melanogaster |
| D. ananassae |
| D. sechellia |
| Other species |
| Trimmed Sequences |
| Translations |
| Multiple Alignments of Protein Sequences |
| Manual Alignment for Poster |
We begin by getting the Ten-m CDS for D. melanogaster from the Gene Record Finder at GEP. The last two segments are here:
>Ten-m:1_536_0 VAFVHDTAGRLETILAGLSSTHYTYQDTTSLVKSVEVQEPGFELRREFKY HAGILKDEKLRFGSKNSLASARYKYAYDGNARLSGIEMAIDDKELPTTRY KYSQNLGQLEVVQDLKITRNAFNRTVIQDSAKQFFAIVDYDQHGRVKSVL MNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNADGHVVEVLG TNNWKYLFDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGF VVKRGEQKYRYNNRGQLIHSFERERFQSWYYYDDRSRLVAWHDNKGNTTQ YYYANPRTPHLVTHVHFPKISRTMKLFYDDRDMLIALEHEDQRYYVATDQ NGSPLAFFDQNGSIVKEMKRTPFGRIIKDTKPEFFVPIDFHGGLIDPHTK LVYTEQRQYDPHVGQWMTPLWETLATEMSHPTDVFIYRYHNNDPINPNKP QNYMIDLDSWLQLFGYDLNNMQSSRYTKLAQYTPQASIKSNTLAPDFGVI SGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRVSYRRGVFGEGVL LSRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVK DNVLKLRDDNEELRRLGGMFNISTHEISDHGGSAAKELRLHGPDAVVIIK YGVDPEQERHRILKHAHKRAVERAWELEKQLVAAGFQGRGDWTEEEKEEL VQHGDVDGWNGIDIHSIHKYPQLADDPGNVAFQRDAKRKRRKTGSSHRSA SNRRQLKFGELSA* >Ten-m:2_602_0 VAFVHDTAGRLETILAGLSSTHYTYQDTTSLVKSVEVQEPGFELRREFKY HAGILKDEKLRFGSKNSLASARYKYAYDGNARLSGIEMAIDDKELPTTRY KYSQNLGQLEVVQDLKITRNAFNRTVIQDSAKQFFAIVDYDQHGRVKSVL MNVKNIDVFRLELDYDLRNRIKSQKTTFGRSTAFDKINYNADGHVVEVLG TNNWKYLFDENGNTVGVVDQGEKFNLGYDIGDRVIKVGDVEFNNYDARGF VVKRGEQKYRYNNRGQLIHSFERERFQSWYYYDDRSRLVAWHDNKGNTTQ YYYANPRTPHLVTHVHFPKISRTMKLFYDDRDMLIALEHEDQRYYVATDQ NGSPLAFFDQNGSIVKEMKRTPFGRIIKDTKPEFFVPIDFHGGLIDPHTK LVYTEQRQYDPHVGQWMTPLWETLATEMSHPTDVFIYRYHNNDPINPNKP QNYMIDLDSWLQLFGYDLNNMQSSRYTKLAQYTPQASIKSNTLAPDFGVI SGLECIVEKTSEKFSDFDFVPKPLLKMEPKMRNLLPRVSYRRGVFGEGVL LSRIGGRALVSVVDGSNSVVQDVVSSVFNNSYFLDLHFSIHDQDVFYFVK DNVLKLRDDNEELRRLGGMFNISTHEISDHGGSAAKELRLHGPDAVVIIK YGVDPEQERHRILKHAHKRAVERAWELEKQLVAAGFQGRGDWTEEEKEEL VQHGDVDGWNGIDIHSIHKYPQLADDPGNVAFQRDAKRKRRKTGSSHRSA SNRRQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGK MEPRDSSEALDEQIL*
If you perform a bl2seq analysis using BLASTP, you will see that they align perfectly. Notice that CDS2 has an internal stop codon. There is a second stop codon beyond that. This is the protein encoded when stop codon readthrough takes place.
The peptide sequence from the first stop to the second is:
*VEAVGADPKQSEWEAGVQANPQLDEIEVADEDDGEQPDYLISVGRKEPKDSSETLEEQIL
We will use the D. melanogaster sequence shown above to retrieve the D. ananassae genomic sequence. For D. ananassae, we will obtain the sequence from the fosmid 1475K17, which contains sequence improved by GEP students at other universities.
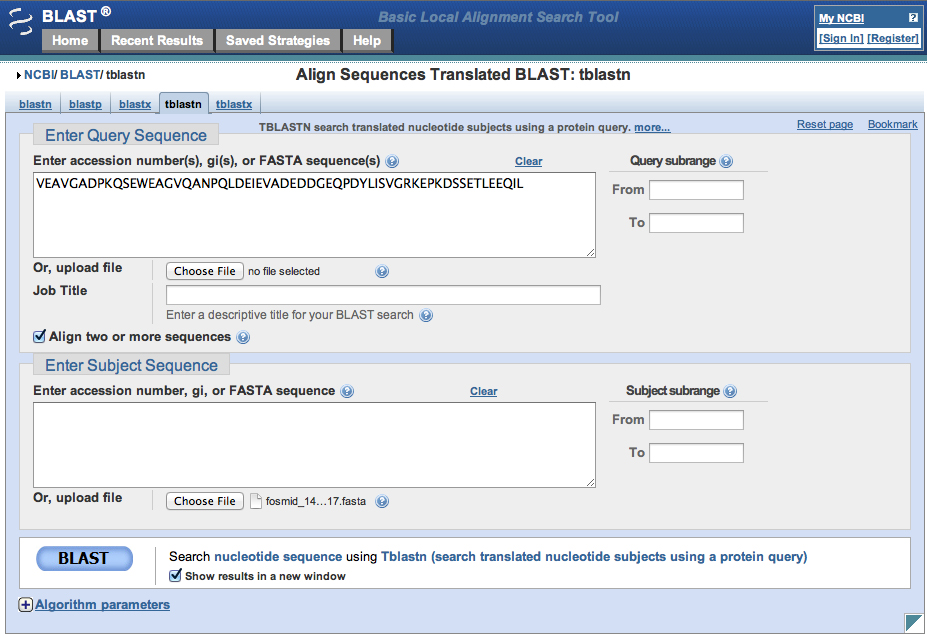
We use bl2seq using TBLASTN to do this. Our query sequence is the peptide above, the subject sequence is the fosmid. This is shown in the screenshots below.

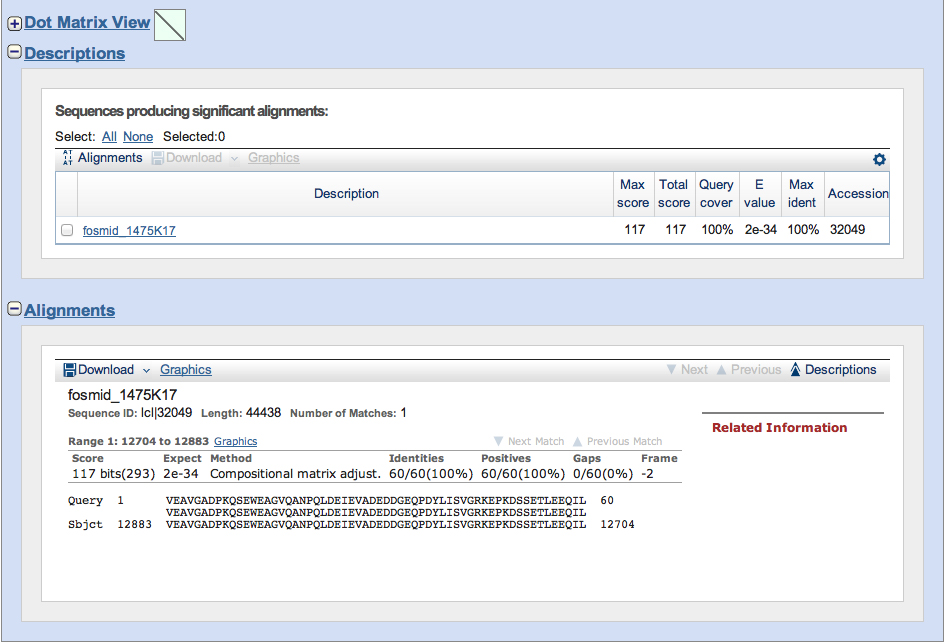
From the alignment, see that the frame is -2, and we get the fosmid coordinates (12883 to 12704). We want to retrieve this nucleotide sequence and the flanking regions.
Go to the GEP UCSC Genome Browser and load fosmid 1475K17, as shown below.
We move in for a closer look, selecting the minus strand. The stop codon in frame -2 that ends the Ten-m-PD and Ten-m-PB isoforms is in view, as is the one that is used in Ten-m-PE when the first stop is read through.
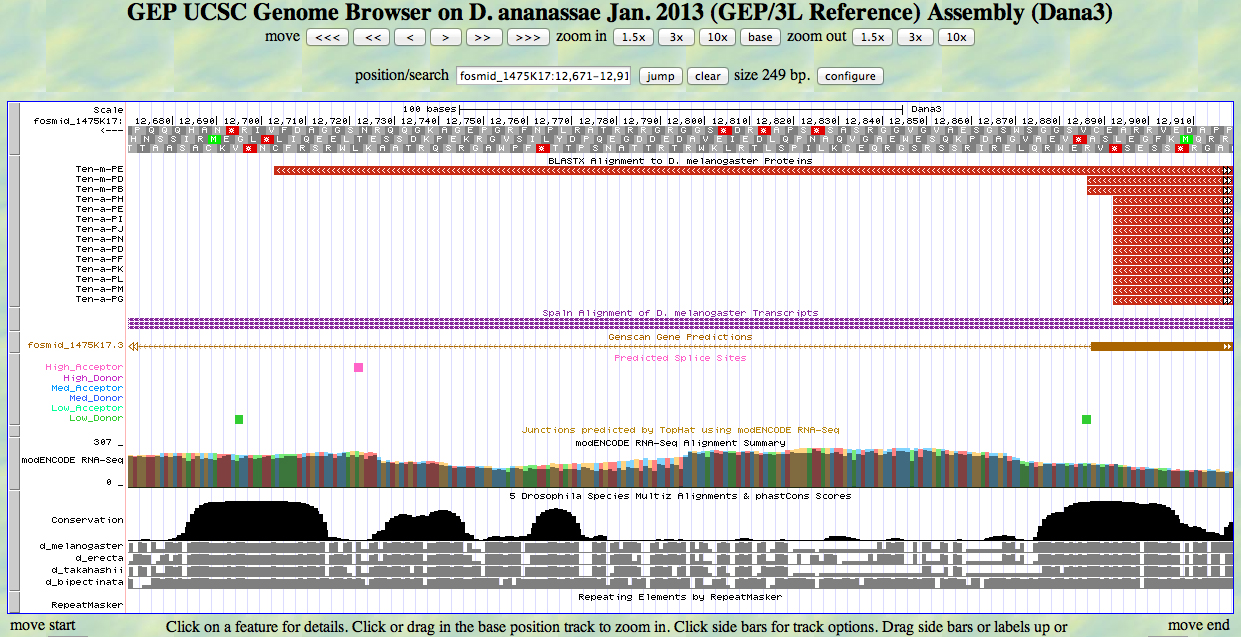
Let's retrieve ten codons upstream of the first stop codon and ten codons downstream of the second stop codon. The first base of the first stop is 12886. Adding 30 bases gives us 12916. The last base of the second stop is 12701. Subtracting 30 bases gives us 12671. Unfortunately, the UCSC Genome Browser at GEP does not have a function to export sequences, so we must do this manually.
To extract the sequence manually, we use a text editor to open the fosmid_1475K17.fasta file. The text is formatted at 50 bases per line, so we need lines 253 (12650/50) through 259 (12950/50). We extract the following sequence:
TCAAGGGCTGAACTGTGACTAATACTAAACTTCACGATTAAAGTTTATAT TAATTGTGTGTTGAATGCCTTTTTTTTAAATGTTTTGTGTGTTTGTGTGT CGCCGTGTTGTGCTGTTGTTGTGGTTGCTGCTGATGCGCATTTCACCTAA TTACAAAATCTGCTCCTCCAGAGTTTCGCTGCTGTCCTTTGGCTCCTTCC GGCCCACGGAAATTAGGTAGTCGGGCTGTTCGCCGTCGTCCTCGTCCGCC ACTTCAATCTCGTCAAGCTGGGGATTAGCTTGCACTCCTGCCTCCCACTC CGACTGCTTCGGATCCGCTCCAACTGCCTCCACTCACGCACTCAGCTCTC
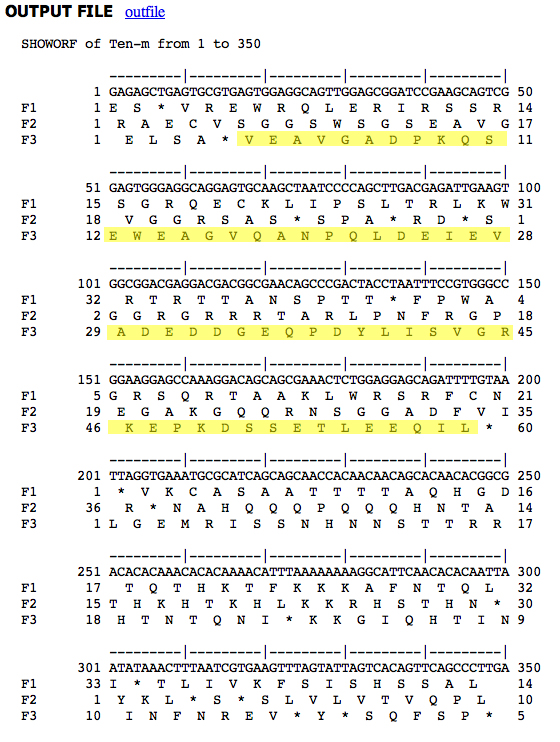
It is a good idea to check this sequence before doing anything else. We go to the EMBOSS toolkit and use the showorf tool. We aren't sure which frame to use, so we select R1, R2, and R3 as shown.

Here is the output, which we search by eye for the peptide sequence.

The peptide sequence is highlighted below.

This is good, but it's no fun reading the sequence backwards, so we use revseq to get the reverse complement as shown.
Here is the output:

We are going to collect a lot of sequences, so we rename this:
>Ten-m D ananassae GAGAGCTGAGTGCGTGAGTGGAGGCAGTTGGAGCGGATCCGAAGCAGTCGGAGTGGGAGG CAGGAGTGCAAGCTAATCCCCAGCTTGACGAGATTGAAGTGGCGGACGAGGACGACGGCG AACAGCCCGACTACCTAATTTCCGTGGGCCGGAAGGAGCCAAAGGACAGCAGCGAAACTC TGGAGGAGCAGATTTTGTAATTAGGTGAAATGCGCATCAGCAGCAACCACAACAACAGCA CAACACGGCGACACACAAACACACAAAACATTTAAAAAAAAGGCATTCAACACACAATTA ATATAAACTTTAATCGTGAAGTTTAGTATTAGTCACAGTTCAGCCCTTGA
This looks good in showorf, as shown.
Frame 3 is the correct frame, as shown.

We didn't get enough sequence before the first stop, so we retrieve lines 253 to 260, obtaining:
TTTTTGAAGCCTGAAAATGAGACAACTGCCTTAAGCCACAGAAAATATTC GAAGCACTTTTCCCAATGCCTAATCGAGGCAAAAATGCATAATTGCATGT GGTGCCCTTACGCAAAAAGGCTGCCTAATTTTTCCTTGAAAAGTGATCAA ATGTATGGCCGACCGGCATTATAATATACCCAGTTATTATTTTAGATTAC GTAAGCACTTTATTCGACATTTTACGAGTAAAAAAATGCAGTGTCTAGAA TACATTAGGGTATGTTTAACAGATTACAGTAAATAAGATTAGCATAATGG TCGACAGTCGGCCGTTGGATAAACTATTGTGTGAAAGCAATTTGGAATTT AGCGACGATTTGCTCACGCTATTGCAACAGATTAAGACAAGCTTAAAGGA CGAGTCCGAGGATCTGTTCTTGCTTGTAGACGCCATGGTAGGCGTTCCGA
The reverse complement is:
>Ten-m D ananassae GACCGGTAACAGCCACCGCAGTGCCTCCAGCCGCCGGCAGATGAAGTTCGGAGAGCTGAG TGCGTGAGTGGAGGCAGTTGGAGCGGATCCGAAGCAGTCGGAGTGGGAGGCAGGAGTGCA AGCTAATCCCCAGCTTGACGAGATTGAAGTGGCGGACGAGGACGACGGCGAACAGCCCGA CTACCTAATTTCCGTGGGCCGGAAGGAGCCAAAGGACAGCAGCGAAACTCTGGAGGAGCA GATTTTGTAATTAGGTGAAATGCGCATCAGCAGCAACCACAACAACAGCACAACACGGCG ACACACAAACACACAAAACATTTAAAAAAAAGGCATTCAACACACAATTAATATAAACTT TAATCGTGAAGTTTAGTATTAGTCACAGTTCAGCCCTTGA
This looks good in showorf:

Now we need the sequence from other species. We start with D. melanogaster. We will use the same method here that is needed for all other species. Go to the FlyBase page for D. melanogaster Ten-m. Enter GBrowse and zoom in on the 3' end as shown in the screenshot below.
Click the button to configure the Decorated FASTA file. This gives the selections shown below. I have selected CDS on a red background and bold for mRNA.

This gives us:
Running this through revseq gives us:
>Ten-m D melanogaster caacgaggagctgcgtcgccttggcggcatgttcaacatatcaacccatgaaatcagcga ccacggaggcagcgccgctaaggagctgcgtcttcacggcccagacgccgttgtcattat caaatatggcgttgatcccgagcaggagcgccaccgcatcctaaagcatgcccacaagcg ggcagtggagcgggcttgggagttggagaagcagctggtggctgccggcttccaaggccg tggcgactggaccgaggaggagaaagaggagctcgtccaacacggtgacgtcgacggctg gaacggaatcgatatccacagcatacacaagtatccgcagctggccgacgaccccggtaa cgtggctttccagcgggacgcgaagcgtaagcgacgcaagaccggcagcagccatcggag tgcctccaaccgccgccagctcaagttcggcgagctgagtgcgtgagtggaggcatatag agagaagctggtggcggagacgaagccagaggagagtgacatgaccgacgaagaatacgg tgaccaggaggactacctaattgccgtgggcaaaatggagccaagggacagcagtgaggc acttgacgagcagattttgtaattaggtgaaatgcgcaacagcgaccacaagaaaaacaa cagcacaactctcgatgacacaacacaaaataagacaacaacaaaaacaaaggaaaagca agcattgaacacactaattaatataaactttaatcatgcttaaagtttagtattagccac aacgcaaccctcgaataaccgaagaaacttcttatgtgtacactggaacagaaatatgta tgttgatgttgagtttaggcgtatgtttagtccatgtgtgtaatgttaaatgtaattttt taaatcagaatcagtgtccccacgggcaatcagcttgcaatggcattttacgccacggtg cgtattatttagattggtccattaaccgattttggtcgag
This looks good in showorf:

We can trim this to:
>Ten-m D melanogaster cgtggctttccagcgggacgcgaagcgtaagcgacgcaagaccggcagcagccatcggag tgcctccaaccgccgccagctcaagttcggcgagctgagtgcgtgagtggaggcatatag agagaagctggtggcggagacgaagccagaggagagtgacatgaccgacgaagaatacgg tgaccaggaggactacctaattgccgtgggcaaaatggagccaagggacagcagtgaggc acttgacgagcagattttgtaattaggtgaaatgcgcaacagcgaccacaagaaaaacaa cagcacaactctcgatgacacaacacaaaataagacaacaacaaaaacaaaggaaaagca
This also looks good in showorf:
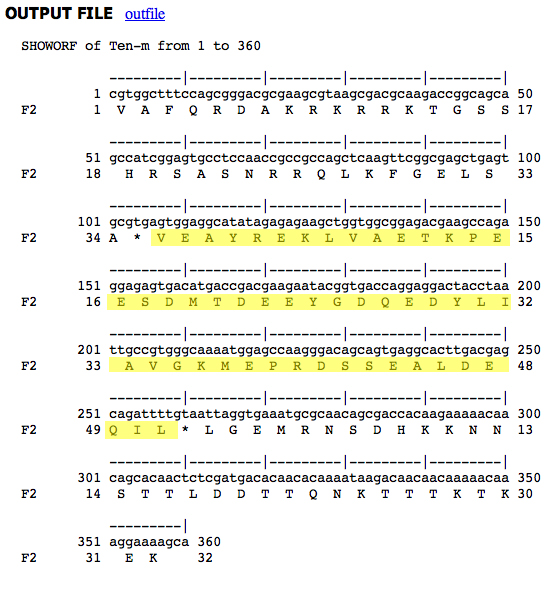
To obtain sequences for other species, we use the AAA orthologs section of the FlyBase page on Ten-m, shown below.
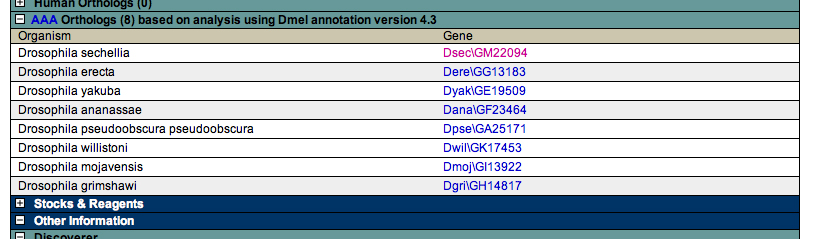
Click the link to go to the ortholog. Here is the view of D. sechellia:
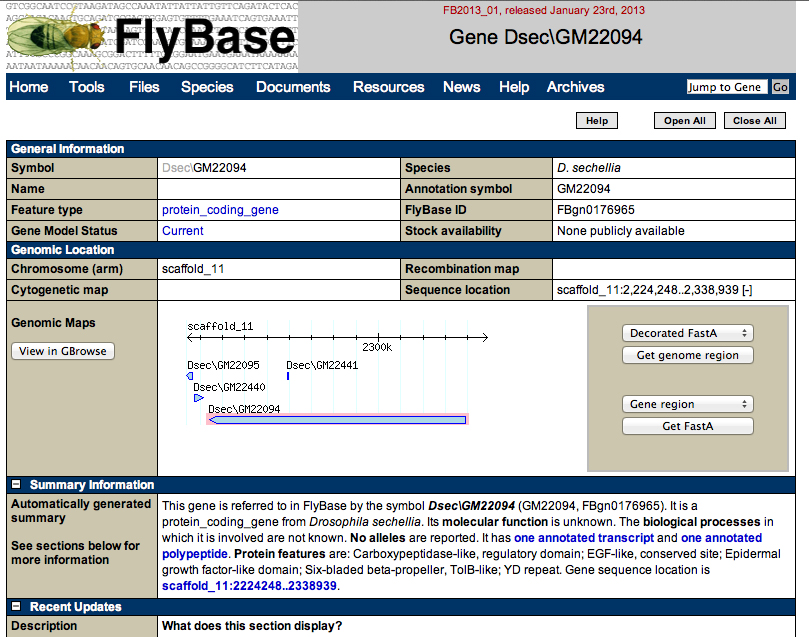
Go to GBrowse as before.
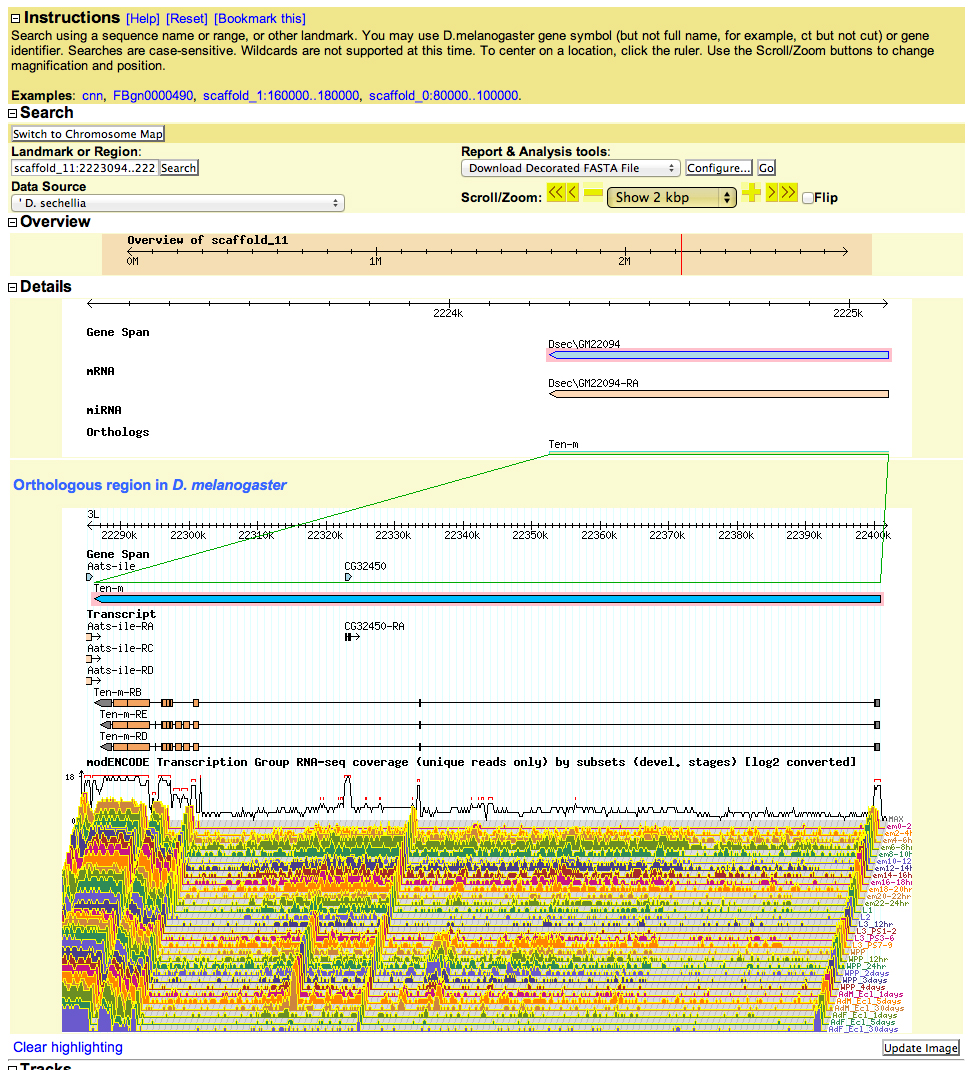
Retrieve sequence:

Here is the raw sequence with a new header, ready to be turned into the reverse complement and trimmed.
>Ten-m D sechellia agaaatatatatcacaatttgtaaaaagtttttaatgcgttttcctttgaataaattcaa cagttaacattataaaatgagtgaaaagagtcgttagagtattgtattgcattgaaaggg gactgataaaaattccttttgattcttggctatccaaattaggtagacgcgtctattgaa tttaaattcgagcattcggcttagagacctattccgctaaaaatagtgttagattaaagg atgcgcgctctttcgtcagtagaggcttgaaaagtattcgttcgtatatacacagagcaa tgaaggagtaggttaagcagcgatcaacattcctgttgccatttgagagtgggctgtgtt ggagtgggccaggtgtggaccaaagactagggagctactatgcagaacttcaatcttttt acatttattcatttcgtagtttaacatctgtacttatcgtatgcatattttgctgtgtaa tactcatcgtttgacatcgtaatgcaactgccttgtttgtggcccggtttgtcctaactc caatttccgaaacaggcgagatcattccgccgttcgtattggttcgatctatgcactctt gtctcgacctaaattggtaatggaccaatctaaataatacgcaccgtggcgtaaaatgcc attgcaagcgggttgcccgtggggacactgattctgatttaaaaaattacattaaacatt acacacatggactaaacatacgcctactgttagcctaaactcaacatcaacatacatatt tctgttccagtgtacacataagaagtttcttcggttattcgagggttgcgttgtggctaa tactaaactttaagcatgattaaagtttatattaattagtgtgttcaatgcttgatgctt gtcctttgttttttttttttttatgtcttattttgtgttgtgtcatcgagagttgtgctg ttgtttttcttgtggtcgctgttgcgcatttcacctaattacaaaatctgctcgtccagt gcctcactgctgtcccttggctcctttttgcccacggcaattaggtagtcctcctggtca ccgtattcttcgtcggtcatgtcactctcctctggctccgcctccgccaccagcttctct ctatacgcctccactcacgcactcagctcgccgaacttgagctggcggcggttggaggca ctccgatggctgctaccggtcttgcgtcgcttccgcttcgcgtcccgctggaaagccacg ttaccggggtcgtcggccagctgcggatacttgtgtatgctgtggatatcgattccgttc cagccgtcgacgtcaccgtgctggacgagctcttctttctcctcctcggtccagtcgcca cgcccttggaagccggcagccaccagctgcttctccaactcccaagcccgctccactgcc cgcttgtgagcatgctttaggatgcggtgtcgctcctgctctggatcaacgccatatttg ataatgacaacggcgtcggggccatgaagtcgcagctccttagcggcgctacctccgtgg tcgctgatttcatgtgttgatatgttgaacatgccgccaaggcgacgcagctcctcgttg tcatcgcgcagtttcaggacattatcctttacgaagtaaaatacgtcttggtcgtggatg ctgaagtgcaggtcaaggaagtatgagttgttgaacacgctgctcactacgtcctgcacc acgctgtttgatccgtcgaccacgctaaccaatgcccgtccgccgattctcgagaggagt acaccttcgccgaacacaccgcgccggtagctgacacgcggcagcaagttgcgcatcttt ggctccattttcagcagcggcttgggcacaaagtcaaagtcactgaacttctcgcttgtt ttttccacgatacactccaggccggagatgacgccaaagtcaggggccaacgtgtttgac ttgatggaggcctgcggcgt
Repeating this approach gives the following raw sequences for other species.
The sequence of D. willistoni and D. mojavensis are already in the reverse orientation; all others are like D. melanogaster and D. ananassae and will need to be reverse-complemented.
>Ten-m D erecta ccaatttaaataatacccaccctgacctaaaatgccactgcaagctgtttgcccgtgggg acactgattctgattcaaaaaattacattaaacattacacacatggactaaacatacgcc tattgtaagcctaaactcaacatcaacatacatatttctgttccagtgttgtgtgtttct tcggttattcgagggctgcgttgtggctaatactaaactttaagcatgattaaagtttgt attaattagtgtattcaatgctggcttttcctttgtatcttgtcttattttgtgttgtgt catcgagagttgtgctgttgtttcggttgtttttgttgtggtcgcatagtttgcgcattt cacctaattacaaaatctgctcgtccagtgcctcactgctgtcccttggctcctttttgc ccacggctattaggtagtcctcctggtcgccatattcctcgtcggtcgtgtcactctcct cgggctctgtctctggctccgcctccaccactagcttctctctatacgcctccactcacg cactcagctcgccgaacttgagctggcgccggttggaggcactccgatggctgctgccgg tcttgcgtcgcttccgcttcgcgtcccgctggaaagcgacgttgccggggtcgtcggcca gttgcggatacttgtgtatgctgtggatatcgattccgttccagccgtcgacgtcgccgt gctggacaagctcctccttctcctcctcggtccagtcgccacgcccttggaagccggcgg ccaccagctgcttctccaattcccaagcacgctccactgcccgtttgtgggcgtgcttta ggatgcggtggcgctcctgctcgggatcaacgccgtattttataatgacaacggcgtcgg gaccgtgcaggcgcaactccttggcggcactgcctccgtgatcgctgatttcatgcgttg atatgttgaacatgccgcccaggcgacgcagctcctcgtt >Ten-m D yakuba tgttttttgtcttattttgtgttgtgtcatcgagagttgtgctgtagtttttgttgtggt cgcttttgcgcatttcacctaattacaaaatctgctcgtccagtgcctcactgctgtccc ttggctcctttttgcccacggctattaggtagtcatcctgatcgccgtattcttcgtcgg tcatgtcactctcctctggctccgcctccaccactagcttttctctaaacgcctccactc acgcactcagttcgccgaacttgagctggcgccggttggaggcactccgatggctgctac cgatcttgcgtcgcttccgtttcgcgtccctctggaaagcgacgttaccgggatcgtcag ccaattgcggatacttgtgtatgctgtggatatcgattccgttccagccgtcgacgtcac cgtgctggacgagctcctccttctcctcctcggtccagtcgccacgcccttggaagccgg cagctaccagctgcttctccaactcccaagcacgctccactgcccgcttgtgagcatgct ttaggatgcgatggcgctcctgctcgggatcaacgccgtatttgataatgacaacggcgt cgggaccgtgcaggcgcaactccttagcggcgctgcctccgtgatcgctgatttcatgcg ttgatatgttgaacatgccgccaaggcgacgcagctcttcgttgtcgtcgcgcagcttca ggacattgtcctttacgaagtaaaatacgtcttggtcgtggatgctgaagtgcaggtcaa ggaagtacgagttgttaaacacgctgctcaccacgtcctggaccacgctgtttgacccgt cgacaacgctaaccaacgcccgtccgccaatcctcgagagaagcacaccttcgccgaata caccacgtcggtaactgacacgcggcagcaagttgcgcatctttggctccattttcagca gcggcttgggcacaaagtcaaagtcactgaacttctcgct >Ten-m D pseudoobscura caaaataggctcgcgatcaaacaatacgagcattgctctgttctattcagttatattttg ttctgttccactccattcggagctctgtccctctgctgatgcattagattcatcaatacg ctcttgtagcgtacatgcggatgccaatgccaatgcaaatgcaaatgcaagcaggctgtt gtccggaaggacactgacttaaattctaaaaaaaaattccagtaaacattacacacatgg actacgcctattgtaagcctaaactcaacatcaacatacacatacatatttctgttcggt tccggtgcacataccagtgccggtgccgttacatatctgggctgggtggcctggtgacta atactaaactttaagcttgatcaaagttcatattaattagtgttttgttgaatgttctct tgtgtgttctcttgtcatgtgtcattgtgctgctgtggttgtggttgtggttggcgctgt tgcgtatttcacctaattacaaaatctgctcctcagccgtctcgctgctgtccctctgct cctttctgcccacggctattaagtagtcctcctgctcgccatactccccatcggccgctt cactctccagctccacatcgagctcgggctcgagctcggtcttgggctcgggctcaagtt gctgctcctctttgtcctcctcgactcacgcattcggctcgccgaccctgagttggcgcc ggctggagctggatctgtggctgcttcctgtcttgcggcgcttgcgcttggcatcgcgct ggaaggccacgttgccgggatcgtcggccagctggggatacttgtgaatgctgtgtatgt caatgccaatccatccatcgacgtcgccgtgttggacgagctcctccttctcctcctcgg tccagtcaccgcgaccctggaagccggcagctaccagctgcttctccagctcccaagccc tctccacggctcgcttgtgggcatgcttgagtatgcgatg >Ten-m D grimshawi attctgtgtgttgttgtgtcgtgatgttttttcttttttttgggtgtgcttggtgcttat cgcctatcgctgctgtatttcacctaagtaattacaaaatctgctcctcggctgcctcgc tgcttgttctgcgaacagccttgcccacagctatcaagtagtcctccggcccgtcatact cttccgttgcttcatctgcagcttcactctccagctcctcctcctccactcacgcgttca gctctccatatttctgctgtcgccgattggagccgatccgatggctgttgccagtcttgc gccgcttacgcttggcatcgcgctggaaggccacatttccgggatcgtctgccagctgtg gatacttgtgtatgctatggatgtcaatgccaatccagccatccacatcaccgtgctgaa tgagctcctctttctcctcctctgtccagtcgccacggccctggaatccggctgcaacca gctgcttctccagctcccaggcacgctcaactgccctcttgtgggcatgcttgagtatgc gatgtcgctcttgctcgggatccacaccgtacttgatgattaccactgcctcggggccat gtaggcgtagctccttagcagcactgccaccgtgatcgctgatctcgtgtgttgatatgt tgaacatgccaccaaggcgacgcagctcctcgttatcgtcgcgcagcttgagaacgttat ccttgacaaagtagaacacatcctgatcatgtatgctgaagtgcagatccaggaagtaag agttgttgaacacagagctaaccacatcctgtacgacactattggagccatccaccacac tgaccagggcacgtccaccaatcctcgagagtagaacaccctctccaaacacggcgcgac gatagctgacacgtggcaatagattgcgcatcttgggctccattttgagcagtggcttgg gtgcaaaatcaaagtcgctaaacttctcgttggtcttctc >Ten-m D willistoni gttctgaagctgcgcgacgataatgaggaactgcgtcgtctgggcggcatgtttaacata tcaacgcacgagattagcgatcatggtggcagtgcggccaaggagttgcgcctccatggc cccgatgcagtggtgatcatcaaatatggtgtagatcccgagcaagagagacaccgcatc ctgaagcatgcccacaaacgggccgtggaacgggcctgggagctggagaagcaattggtg gcagctggcttccaaggacgcggcgactggaccgaagaggagaaggaggaacttgtccag catggcgatgtggacggctggattggcatcgatatacacagcatacacaagtatccccaa ctggccgatgatcctggcaatgtggcctttcagcgggatgccaagcgcaagcgccgcaag accggcaacagtcatcgggtcaactcatcgcgcagacagctcaagttcaccgaattgaat gcgtgagtggagccggacccggacctggataatggggcatacaatgaagaggaagtcgtc ggggatgaggaggatgaagactatggcgaaatggaggactacttgatagccgtgggtaag cagaagcagaagcagcagcagcagcagctatttagcacgagcagcagccgcatcgagcag caggatcagcccaaggagctggtcttgtaattaggtgaaaacaacaacaagaaaaacaac cacaatgatcaacactaattaatataaactttaatcaagcttaaagtttagtattagtat ctaaactctcgagtctgtgtgtttgttgcaattttcctttgactgtgcgcgtctgtacgt tttttttgttgtgtgtactgtttttttttttttttgtattaggcatatgtttagtccatg tgtgtgtaatagttaatgtaaaaccttttaacaaatcgaatgaacaaataagatatatat atatatatatatatatacatagatatatatttgatgctac >Ten-m D mojavensis ttgtcaaggacaacgttctgaagctgcgagatgacaatgaagaattgcgtcgccttggcg gcatgttcaacatatcaacgcacgaaattagcgatcatgggggtagcgctgctaaggagc tgcgccttcatggacccgaagcggtagttataataaagtatggagtggatcccgaacaag agcgacatcgcatactcaagcacgcccacaagcgagcagtagagcgcgcctgggagttgg agaaacagctggtagcagcaggttttcaaggccgtggcgactggacagaagaggaaaaag aggagctcatacaacatggagacgtcgatggctggatcggcattgatattcatagcatac acaagtacccacagctggccgatgatcccggcaatgtggccttccaacgtgatgccaaac gcaagcgccgcaagactggcagcagtcatcacagcagttccagtcggagacagcagaaat atggagaactgaccgcgtgagtggaggaggaccttgagaatgaagctagcgatgaagcca tagaggagtatgacgggccagaggactatttaatagctgtgggcaagttaatccataaga caagcacggatacaaccgttgagcagtttttgtaattacctaggtgaaatgcaacagcga taagaaatataagcgatagacgataaccattaagcacaatacacaaaaatcatgacacga aacacagactttgatcaagtttaaagttgaaaatattattagccaggcgagcccgaagta tcgaacagatcaaccagcaatccacgtaaatctttataaatgtatgtatgttgatgttga ttttaagataacgtaaatagggatactatatgtttagtccacgtgcaacattatatgtac tatattttattggtatctgatgctcgtatgcaaagcggcttcgggcatatagatttgtaa aaaaaattccttgacgaaaaacgcgtcgcttggtgacgta >gnl|dsim|3L:21631222-21631467 type=golden_path_region; loc=3L:1..22553184; ID=3L; dbxref=GB:CM000363; MD5=06629455c1a322db77cad5c0254dfc1b; length=22553184; release=r1.3; species=Dsim; CGCCAGCTCAAGTTCGGCGAGCTGAGTGCGTGAGTGGAGGCGTATAGAGAGAAGCTGGTGGCGGAGGCGGAGCCAGAGGA GAGTGACATGACCGACGAAGAATACGGTGACCAGGAGGACTACCTAATTGCCGTGGGCAAAAAGGAGCCAAGGGACAGCA GTGAGGCACTGGACGAGCAGATTTTGTAATTAGGTGAAATGCGCAACAGCGACCACAAGAAAAACAACAGCACAACTCTC GATGAC
March 25, 2013. We added D. simulans and D. sechellia, reverse-complemented the sequences if necessary, and trimmed them for alignment.
>Ten-m D melanogaster CGCCAGCTCAAGTTCGGCGAGCTGAGTGCGTGAGTGGAGGCATATAG AGAGAAGCTGGTGGCGGAGACGAAGCCAGAGGAGAGTGACATGACCGACGAAGAATACGG TGACCAGGAGGACTACCTAATTGCCGTGGGCAAAATGGAGCCAAGGGACAGCAGTGAGGC ACTTGACGAGCAGATTTTGTAATTAGGTGAAATGCGCAACAGCGACCACAAGAAAAACAA CAGCACAACTCTCGATGACACAA >Ten-m_Dsim CGCCAGCTCAAGTTCGGCGAGCTGAGTGCGTGAGTGGAGGCGTATAGAGAGAAGCTGGTGGCGGAGGCGGAGCCAGAGGA GAGTGACATGACCGACGAAGAATACGGTGACCAGGAGGACTACCTAATTGCCGTGGGCAAAAAGGAGCCAAGGGACAGCA GTGAGGCACTGGACGAGCAGATTTTGTAATTAGGTGAAATGCGCAACAGCGACCACAAGAAAAACAACAGCACAACTCTC GATGAC >Ten-m_Dsec CGCCAGCTCAAGTTCGGCGAGCTGAGTGCGTGAGTGGAGGCGTATAGAGAGAAGCTGGTG GCGGAGGCGGAGCCAGAGGAGAGTGACATGACCGACGAAGAATACGGTGACCAGGAGGAC TACCTAATTGCCGTGGGCAAAAAGGAGCCAAGGGACAGCAGTGAGGCACTGGACGAGCAG ATTTTGTAATTAGGTGAAATGCGCAACAGCGACCACAAGAAAAACAACAGCACAACTCTC GATGAC >Ten-m D erecta CGCCAGCTCAAGTTCGGCGAGCTGAGTGCGTGAGTGGAGGCGTATAGA GAGAAGCTAGTGGTGGAGGCGGAGCCAGAGACAGAGCCCGAGGAGAGTGACACGACCGAC GAGGAATATGGCGACCAGGAGGACTACCTAATAGCCGTGGGCAAAAAGGAGCCAAGGGAC AGCAGTGAGGCACTGGACGAGCAGATTTTGTAATTAGGTGAAATGCGCAAACTATGCGAC CACAACAAAAACAACCGAAACA >Ten-m D ananassae CGGCAGATGAAGTTCGGAGAGCTGAG TGCGTGAGTGGAGGCAGTTGGAGCGGATCCGAAGCAGTCGGAGTGGGAGGCAGGAGTGCA AGCTAATCCCCAGCTTGACGAGATTGAAGTGGCGGACGAGGACGACGGCGAACAGCCCGA CTACCTAATTTCCGTGGGCCGGAAGGAGCCAAAGGACAGCAGCGAAACTCTGGAGGAGCA GATTTTGTAATTAGGTGAAATGCGCATCAGCAGCAACCACAACA >Ten-m D yakuba CGCCAGCTCAAGTTCGGCGAACTGAGTGCGTGAGTGGAGGCGTTTAGAGAA AAGCTAGTGGTGGAGGCGGAGCCAGAGGAGAGTGACATGACCGACGAAGAATACGGCGAT CAGGATGACTACCTAATAGCCGTGGGCAAAAAGGAGCCAAGGGACAGCAGTGAGGCACTG GACGAGCAGATTTTGTAATTAGGTGAAATGCGCAAAAGCGACCACAACAAAAACTACAGC ACAACTCTCGATGACACAA >Ten-m D pseudoobscura CGCCAACTCAGGGTCGGC GAGCCGAATGCGTGAGTCGAGGAGGACAAAGAGGAGCAGCAACTTGAGCCCGAGCCCAAG ACCGAGCTCGAGCCCGAGCTCGATGTGGAGCTGGAGAGTGAAGCGGCCGATGGGGAGTAT GGCGAGCAGGAGGACTACTTAATAGCCGTGGGCAGAAAGGAGCAGAGGGACAGCAGCGAG ACGGCTGAGGAGCAGATTTTGTAATTAGGTGAAATACGCAACAGCGCCAACC >Ten-m D willistoni AGACAGCTCAAGTTCACCGAATTGAAT GCGTGAGTGGAGCCGGACCCGGACCTGGATAATGGGGCATACAATGAAGAGGAAGTCGTC GGGGATGAGGAGGATGAAGACTATGGCGAAATGGAGGACTACTTGATAGCCGTGGGTAAG CAGAAGCAGAAGCAGCAGCAGCAGCAGCTATTTAGCACGAGCAGCAGCCGCATCGAGCAG CAGGATCAGCCCAAGGAGCTGGTCTTGTAATTAGGTGAAAACA >Ten-m D mojavensis AGACAGCAGAAAT ATGGAGAACTGACCGCGTGAGTGGAGGAGGACCTTGAGAATGAAGCTAGCGATGAAGCCA TAGAGGAGTATGACGGGCCAGAGGACTATTTAATAGCTGTGGGCAAGTTAATCCATAAGA CAAGCACGGATACAACCGTTGAGCAGTTTTTGTAATTACCTAGGTGAAATGCAACAGCGA TAAGAAATATAAGCGATAGACGATAACCATTAAGCACAATACACAAAAATCATGACA >Ten-m D grimshawi CGACAGCAGAAATATGGAGAGCTGAACGCGTGAGTGGAGGAG GAGGAGCTGGAGAGTGAAGCTGCAGATGAAGCAACGGAAGAGTATGACGGGCCGGAGGAC TACTTGATAGCTGTGGGCAAGGCTGTTCGCAGAACAAGCAGCGAGGCAGCCGAGGAGCAG ATTTTGTAATTACTTAGGTGAAATACAGCAGCGATAGGCGATAAGCACCAAGCACACCCA AAAAAAAGAA
>Ten-m_Dmel RQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQ IL*LGEMRNSDHKKNNSTTLDDTX >Ten-m_Dsim RQLKFGELSA*VEAYREKLVAEAEPEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQ IL*LGEMRNSDHKKNNSTTLDD >Ten-m_Dsec RQLKFGELSA*VEAYREKLVAEAEPEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQ IL*LGEMRNSDHKKNNSTTLDD >Ten-m_Dyak RQLKFGELSA*VEAFREKLVVEAEPEESDMTDEEYGDQDDYLIAVGKKEPRDSSEALDEQ IL*LGEMRKSDHNKNYSTTLDDTX >Ten-m_Dere RQLKFGELSA*VEAYREKLVVEAEPETEPEESDTTDEEYGDQEDYLIAVGKKEPRDSSEA LDEQIL*LGEMRKLCDHNKNNRNX >Ten-m_Dana RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQLDEIEVADEDDGEQPDYLISVGRKEPK DSSETLEEQIL*LGEMRISSNHNX >Ten-m_Dmoj RQQKYGELTA*VEEDLENEASDEAIEEYDGPEDYLIAVGKLIHKTSTDTTVEQFL*LPR* NATAIRNISDRR*PLSTIHKNHDX >Ten-m_Dgri RQQKYGELNA*VEEEELESEAADEATEEYDGPEDYLIAVGKAVRRTSSEAAEEQIL*LLR *NTAAIGDKHQAHPKKRX >Ten-m_Dpse RQLRVGEPNA*VEEDKEEQQLEPEPKTELEPELDVELESEAADGEYGEQEDYLIAVGRKE QRDSSETAEEQIL*LGEIRNSANX >Ten-m_Dwil RQLKFTELNA*VEPDPDLDNGAYNEEEVVGDEEDEDYGEMEDYLIAVGKQKQKQQQQQLF STSSSRIEQQDQPKELVL*LGENX
CLUSTAL O(1.1.0) multiple sequence alignment
Ten-m_Dmel RQLKFGELSA*VEAYREKLV-AETK----P-------EESDMTDEEYGDQEDYLIAVGKM
Ten-m_Dsim RQLKFGELSA*VEAYREKLV-AEAE----P-------EESDMTDEEYGDQEDYLIAVGKK
Ten-m_Dsec RQLKFGELSA*VEAYREKLV-AEAE----P-------EESDMTDEEYGDQEDYLIAVGKK
Ten-m_Dere RQLKFGELSA*VEAYREKLV-VEAEPETEP-------EESDTTDEEYGDQEDYLIAVGKK
Ten-m_Dana RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQL---DEIEVADEDDGEQPDYLISVGRK
Ten-m_Dyak RQLKFGELSA*VEAFREKLV-VEAE----P-------EESDMTDEEYGDQDDYLIAVGKK
Ten-m_Dpse RQLRVGEPNA*VEEDKEEQQ-LEPEPKTELEPELDVELESEAADGEYGEQEDYLIAVGRK
Ten-m_Dmoj RQQKYGELTA*VE-EDLEN------------------EASDEAIEEYDGPEDYLIAVGKL
Ten-m_Dgri RQQKYGELNA*VEEEELES------------------EAADEATEEYDGPEDYLIAVGKA
Ten-m_Dwil RQLKFTELNA*VEPDPDLDN----------GAYNEEEVVGDEEDEDYGEMEDYLIAVGKQ
** : * .**** : : ****:**:
Ten-m_Dmel EPRDSSEAL--------------DEQIL*LGEMRNSD-HKKNNSTTLDDTX------
Ten-m_Dsim EPRDSSEAL--------------DEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dsec EPRDSSEAL--------------DEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dere EPRDSSEAL--------------DEQIL*LGEMRKLCDHNKNNRNX-----------
Ten-m_Dana EPKDSSETL--------------EEQIL*LGEMRISSNHNX----------------
Ten-m_Dyak EPRDSSEAL--------------DEQIL*LGEMRKSD-HNKNYSTTLDDTX------
Ten-m_Dpse EQRDSSETA--------------EEQIL*LGEIRNSANX------------------
Ten-m_Dmoj IHKTSTDTT--------------VEQFL*LPR*NATAIRNISDRR*PLSTIHKNHDX
Ten-m_Dgri VRRTSSEAA--------------EEQIL*LLR*NTAAIGDKHQAHPKKRX-------
Ten-m_Dwil KQKQQQQQLFSTSSSRIEQQDQPKELVL*LGENX-----------------------
: . : * .*** .
CLUSTAL O(1.1.0) multiple sequence alignment
Ten-m_Dmel RQLKFGELSA*VEAYREKLV-AETK----P-------EESDMTDEEYGDQEDYLIAVGKM
Ten-m_Dsim RQLKFGELSA*VEAYREKLV-AEAE----P-------EESDMTDEEYGDQEDYLIAVGKK
Ten-m_Dsec RQLKFGELSA*VEAYREKLV-AEAE----P-------EESDMTDEEYGDQEDYLIAVGKK
Ten-m_Dyak RQLKFGELSA*VEAFREKLV-VEAE----P-------EESDMTDEEYGDQDDYLIAVGKK
Ten-m_Dere RQLKFGELSA*VEAYREKLV-VEAEPETEP-------EESDTTDEEYGDQEDYLIAVGKK
Ten-m_Dana RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQL---DEIEVADEDDGEQPDYLISVGRK
Ten-m_Dmoj RQQKYGELTA*VE-EDLEN------------------EASDEAIEEYDGPEDYLIAVGKL
Ten-m_Dgri RQQKYGELNA*VEEEELES------------------EAADEATEEYDGPEDYLIAVGKA
Ten-m_Dpse RQLRVGEPNA*VEEDKEEQQ-LEPEPKTELEPELDVELESEAADGEYGEQEDYLIAVGRK
** : ** .**** . : : : ****:**:
Ten-m_Dmel EPRDSSEALDEQIL*LGEMRNSD-HKKNNSTTLDDTX------
Ten-m_Dsim EPRDSSEALDEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dsec EPRDSSEALDEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dyak EPRDSSEALDEQIL*LGEMRKSD-HNKNYSTTLDDTX------
Ten-m_Dere EPRDSSEALDEQIL*LGEMRKLCDHNKNNRNX-----------
Ten-m_Dana EPKDSSETLEEQIL*LGEMRISSNHNX----------------
Ten-m_Dmoj IHKTSTDTTVEQFL*LPR*NATAIRNISDRR*PLSTIHKNHDX
Ten-m_Dgri VRRTSSEAAEEQIL*LLR*NTAAIGDKHQAHPKKRX-------
Ten-m_Dpse EQRDSSETAEEQIL*LGEIRNSANX------------------
: *::: **:*** . .
CLUSTAL O(1.1.0) multiple sequence alignment
Ten-m_Dmel RQLKFGELSA*VEAYREKLV-----AETK----PEESDMTDEEYGDQEDYLIAVGKMEPR
Ten-m_Dsim RQLKFGELSA*VEAYREKLV-----AEAE----PEESDMTDEEYGDQEDYLIAVGKKEPR
Ten-m_Dsec RQLKFGELSA*VEAYREKLV-----AEAE----PEESDMTDEEYGDQEDYLIAVGKKEPR
Ten-m_Dyak RQLKFGELSA*VEAFREKLV-----VEAE----PEESDMTDEEYGDQDDYLIAVGKKEPR
Ten-m_Dere RQLKFGELSA*VEAYREKLV-----VEAEPETEPEESDTTDEEYGDQEDYLIAVGKKEPR
Ten-m_Dana RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQLDEIEVADEDDGEQPDYLISVGRKEPK
Ten-m_Dmoj RQQKYGELTA*VE-ED---------------LENEASDEAIEEYDGPEDYLIAVGKLIHK
Ten-m_Dgri RQQKYGELNA*VEEEE---------------LESEAADEATEEYDGPEDYLIAVGKAVRR
** *:***.**** : : : *: ****:**: :
Ten-m_Dmel DSSEALDEQIL*LGEMRNSD-HKKNNSTTLDDTX------
Ten-m_Dsim DSSEALDEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dsec DSSEALDEQIL*LGEMRNSD-HKKNNSTTLDD--------
Ten-m_Dyak DSSEALDEQIL*LGEMRKSD-HNKNYSTTLDDTX------
Ten-m_Dere DSSEALDEQIL*LGEMRKLCDHNKNNRNX-----------
Ten-m_Dana DSSETLEEQIL*LGEMRISSNHNX----------------
Ten-m_Dmoj TSTDTTVEQFL*LPR*NATAIRNISDRR*PLSTIHKNHDX
Ten-m_Dgri TSSEAAEEQIL*LLR*NTAAIGDKHQAHPKKRX-------
*::: **:*** . . .
>Dmel_Ten-m RQLKFGELSA*VEAYREKLV-----AETK----PEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQIL*LGEMRNSDHK >Dsim_Ten-m RQLKFGELSA*VEAYREKLV-----AEAE----PEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQIL*LGEMRNSDHK >Dsec_Ten-m RQLKFGELSA*VEAYREKLV-----AEAE----PEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQIL*LGEMRNSDHK >Dyak_Ten-m RQLKFGELSA*VEAFREKLV-----VEAE----PEESDMTDEEYGDQDDYLIAVGKKEPRDSSEALDEQIL*LGEMRKSDHN >Dere_Ten-m RQLKFGELSA*VEAYREKLV-----VEAEPETEPEESDTTDEEYGDQEDYLIAVGKKEPRDSSEALDEQIL*LGEMRKLCDH >Dana_Ten-m RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQLDEIEVADEDDGEQPDYLISVGRKEPKDSSETLEEQIL*LGEMRISSNH >Dmoj_Ten-m RQQKYGELTA*VE-ED---------------LENEASDEAIEEYDGPEDYLIAVGKLIHKTSTDTTVEQFL*LPR*NATAIR >Dgri_Ten-m RQQKYGELNA*VEEEE---------------LESEAADEATEEYDGPEDYLIAVGKAVRRTSSEAAEEQIL*LLR*NTAAIG >Dmel_aa R Q L K F G E L S A * V E A Y R E K L V - - - - - A E T K - - - - P E E S D M T D E E Y G D Q E D Y L I A V G K M E P R D S S E A L D E Q I L * L G E M R N S D H K >Dmel_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCA TAT AGA GAG AAG CTG GTG - - - - - GCG GAG ACG AAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA ATG GAG CCA AGG GAC AGC AGT GAG GCA CTT GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG AAA AAC AAC AGC ACAACTCTCGATGACACAA >Dsim_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTG GTG - - - - - GCG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG AAA AAC AAC AGC ACAACTCTCGATGAC >Dsec_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTG GTG - - - - - GCG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG AAA AAC AAC AGC ACAACTCTCGATGAC >Dyak_Ten-m CGC CAG CTC AAG TTC GGC GAA CTG AGT GCG TGA GTG GAG GCG TTT AGA GAA AAG CTA GTG - - - - - GTG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC GAA GAA TAC GGC GAT CAG GAT GAC TAC CTA ATA GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAA AGC GAC CAC AAC AAAAACTACAGCACAACTCTCGATGACACAA >Dere_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTA GTG - - - - - GTG GAG GCG GAG CCA GAG ACA GAG CCC GAG GAG AGT GAC ACG ACC GAC GAG GAA TAT GGC GAC CAG GAG GAC TAC CTA ATA GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAA CTA TGC GAC CAC AACAAAAACAACCGAAACA >Dana_Ten-m CGG CAG ATG AAG TTC GGA GAG CTG AGT GCG TGA GTG GAG GCA GTT GGA GCG GAT CCG AAG CAG TCG GAG TGG GAG GCA GGA GTG CAA GCT AAT CCC CAG CTT GAC GAG ATT GAA GTG GCG GAC GAG GAC GAC GGC GAA CAG CCC GAC TAC CTA ATT TCC GTG GGC CGG AAG GAG CCA AAG GAC AGC AGC GAA ACT CTG GAG GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC ATC AGC AGC AAC CAC AACA >Dmoj_Ten-m AGA CAG CAG AAA TAT GGA GAA CTG ACC GCG TGA GTG GAG - GAG GAC - - - - - - - - - - - - - - - CTT GAG AAT GAA GCT AGC GAT GAA GCC ATA GAG GAG TAT GAC GGG CCA GAG GAC TAT TTA ATA GCT GTG GGC AAG TTA ATC CAT AAG ACA AGC ACG GAT ACA ACC GTT GAG CAG TTT TTG TAA TTA CCT AGG TGA AAT GCA ACA GCG ATA AGA AATATAAGCGATAGACGATAACCATTAAGCACAATACACAAAAATCATGACA >Dgri_Ten-m CGA CAG CAG AAA TAT GGA GAG CTG AAC GCG TGA GTG GAG GAG GAG GAG - - - - - - - - - - - - - - - CTG GAG AGT GAA GCT GCA GAT GAA GCA ACG GAA GAG TAT GAC GGG CCG GAG GAC TAC TTG ATA GCT GTG GGC AAG GCT GTT CGC AGA ACA AGC AGC GAG GCA GCC GAG GAG CAG ATT TTG TAA TTA CTT AGG TGA AAT ACA GCA GCG ATA GGC GATAAGCACCAAGCACACCCAAAAAAAAGAA >Dmel_aa R Q L K F G E L S A * V E A Y R E K L V - - - - - A E T K - - - - P E E S D M T D >Dmel_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCA TAT AGA GAG AAG CTG GTG - - - - - GCG GAG ACG AAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC >Dsim_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTG GTG - - - - - GCG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC >Dsec_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTG GTG - - - - - GCG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC >Dyak_Ten-m CGC CAG CTC AAG TTC GGC GAA CTG AGT GCG TGA GTG GAG GCG TTT AGA GAA AAG CTA GTG - - - - - GTG GAG GCG GAG - - - - CCA GAG GAG AGT GAC ATG ACC GAC >Dere_Ten-m CGC CAG CTC AAG TTC GGC GAG CTG AGT GCG TGA GTG GAG GCG TAT AGA GAG AAG CTA GTG - - - - - GTG GAG GCG GAG CCA GAG ACA GAG CCC GAG GAG AGT GAC ACG ACC GAC >Dana_Ten-m CGG CAG ATG AAG TTC GGA GAG CTG AGT GCG TGA GTG GAG GCA GTT GGA GCG GAT CCG AAG CAG TCG GAG TGG GAG GCA GGA GTG CAA GCT AAT CCC CAG CTT GAC GAG ATT GAA GTG GCG GAC >Dmoj_Ten-m AGA CAG CAG AAA TAT GGA GAA CTG ACC GCG TGA GTG GAG - GAG GAC - - - - - - - - - - - - - - - CTT GAG AAT GAA GCT AGC GAT GAA GCC ATA >Dgri_Ten-m CGA CAG CAG AAA TAT GGA GAG CTG AAC GCG TGA GTG GAG GAG GAG GAG - - - - - - - - - - - - - - - CTG GAG AGT GAA GCT GCA GAT GAA GCA ACG >Dmel_aa E E Y G D Q E D Y L I A V G K M E P R D S S E A L D E Q I L * L G E M R N S D H K >Dmel_Ten-m GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA ATG GAG CCA AGG GAC AGC AGT GAG GCA CTT GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG >Dsim_Ten-m GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG >Dsec_Ten-m GAA GAA TAC GGT GAC CAG GAG GAC TAC CTA ATT GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAC AGC GAC CAC AAG >Dyak_Ten-m GAA GAA TAC GGC GAT CAG GAT GAC TAC CTA ATA GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAA AGC GAC CAC AAC >Dere_Ten-m GAG GAA TAT GGC GAC CAG GAG GAC TAC CTA ATA GCC GTG GGC AAA AAG GAG CCA AGG GAC AGC AGT GAG GCA CTG GAC GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC AAA CTA TGC GAC CAC >Dana_Ten-m GAG GAC GAC GGC GAA CAG CCC GAC TAC CTA ATT TCC GTG GGC CGG AAG GAG CCA AAG GAC AGC AGC GAA ACT CTG GAG GAG CAG ATT TTG TAA TTA GGT GAA ATG CGC ATC AGC AGC AAC CAC >Dmoj_Ten-m GAG GAG TAT GAC GGG CCA GAG GAC TAT TTA ATA GCT GTG GGC AAG TTA ATC CAT AAG ACA AGC ACG GAT ACA ACC GTT GAG CAG TTT TTG TAA TTA CCT AGG TGA AAT GCA ACA GCG ATA AGA >Dgri_Ten-m GAA GAG TAT GAC GGG CCG GAG GAC TAC TTG ATA GCT GTG GGC AAG GCT GTT CGC AGA ACA AGC AGC GAG GCA GCC GAG GAG CAG ATT TTG TAA TTA CTT AGG TGA AAT ACA GCA GCG ATA GGC
Calls of identity, synonymous, conservative, nonconservative were made on the alignment by hand. Here is a set of checks using bl2seq.
Dmel (query) vs. Dsim (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQ 60
RQLKFGELSA*VEAYREKLVAE +PEESDMTDEEYGDQEDYLIAVGK EPRDSSEALDEQ
Sbjct 1 RQLKFGELSA*VEAYREKLVAEAEPEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQ 60
Query 61 IL*LGEMRNSDHKKNNSTTLDD 82
IL*LGEMRNSDHKKNNSTTLDD
Sbjct 61 IL*LGEMRNSDHKKNNSTTLDD 82
Dmel (query) vs. Dsec (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQ 60
RQLKFGELSA*VEAYREKLVAE +PEESDMTDEEYGDQEDYLIAVGK EPRDSSEALDEQ
Sbjct 1 RQLKFGELSA*VEAYREKLVAEAEPEESDMTDEEYGDQEDYLIAVGKKEPRDSSEALDEQ 60
Query 61 IL*LGEMRNSDHKKNNSTTLDD 82
IL*LGEMRNSDHKKNNSTTLDD
Sbjct 61 IL*LGEMRNSDHKKNNSTTLDD 82
Dmel (query) vs. Dyak (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQ 60
RQLKFGELSA*VEA+REKLV E +PEESDMTDEEYGDQ+DYLIAVGK EPRDSSEALDEQ
Sbjct 1 RQLKFGELSA*VEAFREKLVVEAEPEESDMTDEEYGDQDDYLIAVGKKEPRDSSEALDEQ 60
Query 61 IL*LGEMRNSDHKKNNSTTLDDT 83
IL*LGEMR SDH KN STTLDDT
Sbjct 61 IL*LGEMRKSDHNKNYSTTLDDT 83
Dmel (query) vs. Dere (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPEE----SDMTDEEYGDQEDYLIAVGKMEPRDSSEA 56
RQLKFGELSA*VEAYREKLV E +PE SD TDEEYGDQEDYLIAVGK EPRDSSEA
Sbjct 1 RQLKFGELSA*VEAYREKLVVEAEPETEPEESDTTDEEYGDQEDYLIAVGKKEPRDSSEA 60
Query 57 LDEQIL*LGEMRN-SDHKKNN 76
LDEQIL*LGEMR DH KNN
Sbjct 61 LDEQIL*LGEMRKLCDHNKNN 81
Dmel (query) vs. Dana (subject)
Query 1 RQLKFGELSA*VEA---------YREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPR 51
RQ+KFGELSA*VEA + + A + +E ++ DE+ G+Q DYLI+VG+ EP+
Sbjct 1 RQMKFGELSA*VEAVGADPKQSEWEAGVQANPQLDEIEVADEDDGEQPDYLISVGRKEPK 60
Query 52 DSSEALDEQIL*LGEMR-NSDH 72
DSSE L+EQIL*LGEMR +S+H
Sbjct 61 DSSETLEEQIL*LGEMRISSNH 82
Dmel (query) vs. Dmoj (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPEESDMTDEEYGDQEDYLIAVGKMEPRDSSEALDEQ 60
RQ K+GEL+A*VE E E SD EEY EDYLIAVGK+ + S++ EQ
Sbjct 1 RQQKYGELTA*VEEDLEN-------EASDEAIEEYDGPEDYLIAVGKLIHKTSTDTTVEQ 53
Query 61 IL*L 64
L*L
Sbjct 54 FL*L 57
Dmel (query) vs. Dgri (subject)
Query 33 EEYGDQEDYLIAVGKMEPRDSSEALDEQIL*L 64
EEY EDYLIAVGK R SSEA +EQIL*L
Sbjct 27 EEYDGPEDYLIAVGKAVRRTSSEAAEEQIL*L 58
Dmel (query) vs. Dpse (subject)
Query 1 RQLKFGELSA*VEAYREKLVAETKPE-----------ESDMTDEEYGDQEDYLIAVGKME 49
RQL+ GE +A*VE +E+ E +P+ ES+ D EYG+QEDYLIAVG+ E
Sbjct 1 RQLRVGEPNA*VEEDKEEQQLEPEPKTELEPELDVELESEAADGEYGEQEDYLIAVGRKE 60
Query 50 PRDSSEALDEQIL*LGEMRNS 70
RDSSE +EQIL*LGE+RNS
Sbjct 61 QRDSSETAEEQIL*LGEIRNS 81
Dmel (query) vs. Dwil (subject)
Query 1 RQLKFGELSA*VE--------AYREKLVAETKPEESDMTDEEYGDQEDYLIAVG 46
RQLKF EL+A*VE AY E+ V + DE+YG+ EDYLIAVG
Sbjct 1 RQLKFTELNA*VEPDPDLDNGAYNEEEVVGDE------EDEDYGEMEDYLIAVG 48