 |
|||||||||||||
|
 |
|||||||||||||
|
1. Starting Project Title: Using Metagenomics and Next Generation Sequencing Technologies as a First Step to characterize New Mexican algae communities seeking the best populations for algae biodiesel production. The development of a workable testing matrix for algae metagenomic populations using (2-3) community samples plied against (3-4) culling challenges (i.e. high light intensity, high temperature, high salinity and cold temperature) with the expected result that the survivor(s) would be the best candidate(s) and promoted to the next testing step.
2. Statement of the Problem Algae biofuels is one most promising renewable energy source and has begun supplying the world with biodiesel. Currently the biofuel industry uses single specie algae cultures to create biodiesel, but in nature algae exist in communities. It is algae in communities that may have an advantage for the creation of algae biodiesel. Unfortunately there has been little if any research investigating how algae communities interact much less understanding the larger picture of how algae populations can benefit biodiesel production. It remains to be discovered whether a single specie or the group algae culture has the most benefits for the production of algae biodiesel. The proposed research seeks to determine what algae species are in New Mexico metagenomic samples and how these algae populations can adapt to group culturing with the specific aim to produce maximum biodiesel.
Sampling uncultured environmental algae populations (called metagenomic sampling) from New Mexico springs requires immediate genomic analysis because many algae species cannot be cultured in vitro. The analysis of all the algae population genomics will answer the question of who is there and what they may be doing. Sequencing an entire algae population requires newer sequencing technologies called Next Generation Sequencing (NGS). The Roche 454 GS FLX pyrosequencer (NGS) can run an entire population of organisms and sequence their genomes in one run delivering raw data and sequences for the entire population. The data can be compared to online genetic databases for identification. NGS requires extensive use of biocomputing and bioinformatics which has been expanding capacities and speed to handle the large volumes of genomic data from NGS. Up until this point NGS has been applied to bacterial populations and the application to algae is entirely new.
The world runs on energy and the primary energy in the United States (US) is crude petroleum oil. The crude oil supply is finite and will be exhausted one day. Creating a renewable sustainable energy source in algae biofuel will start weaning the US off foreign oil and slow the depletion of US crude oil reserves. Algae biofuels as a living renewable oil source offers a large energy opportunity and the industrial infrastructure to put algae biofuel into production is still in its infancy. Algae biofuel production primarily makes biodiesel and the algae biodiesel cycle is shown in Figure 1.
United States Crude Oil Consumption
The world transportation systems have run on crude petroleum oil for over a century and for 2012 the United States consumes 18.2 million barrels of oil daily. (Zhou 2012) The crude petroleum production in the United States has increased 7.3% in the last year and was the result of 50 more oil rigs put into service. The technical complexity to remove oil from the ground has increased dramatically and the cost to produce a barrel of oil has risen substantially from the past where oil was extracted by direct drilling and pumping. A technology called horizontal fracturing (nicknamed fracking) which has added additional US crude oil sources. Unfortunately fracking has caused considerable environmental controversy due to its potential to contaminate ground water aquifers. Increasing world oil consumption by China, Europe, India and other countries creates competition for foreign oil and adding a renewable oil source from algae adds a new domestic source.
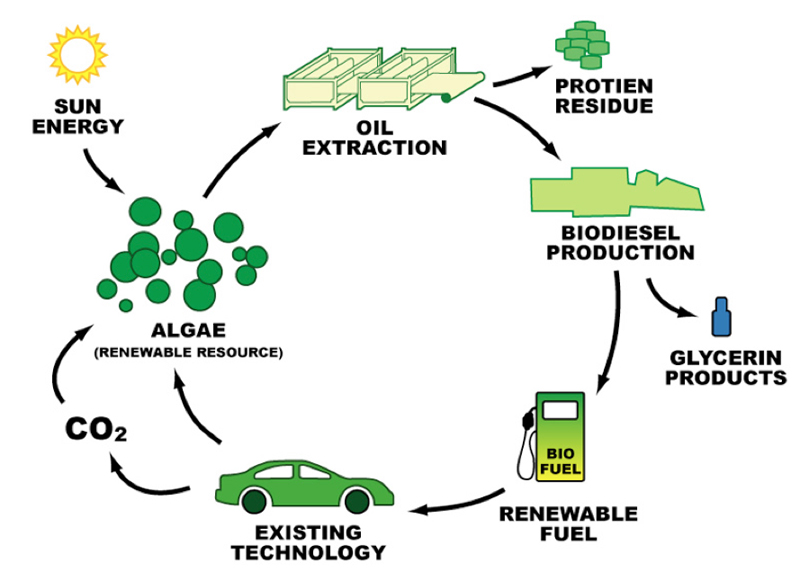
Figure 1 - Algae Biodiesel Cycle, Christopher Gully (2012) Fuel from Slime: Suspension of Disbelief, Carbon Talks, http://blog.carbontalks.ca/archives/700/ trackback.
3. Applicable Prior Algae Research
A Look Back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from Algae (Sheehan 1998)
In the landmark Aquatic Species Program (1978 to 1996) conducted by the Department of Energy Office of Fuels and Development research effort investigated the possibility that high lipid content algae could produce biodiesel. (Sheehan 1998) The Aquatic Species Program looked a large range of algae species and wanted to find algae capable of growth under severe conditions such as extremes of temperature, pH and salinity. All species involved in the research were involved as single species.
The program researchers built up a collection of 300 species winnowed down from the initial 3000 species. Algae were stressed by nitrogen starvation which caused the algae cells to sequester lipids (also know as triacylglycerol, TAGs) in their cells at the expense of building proteins for cell growth. The program also genetically modified algae with the program's newly discovered Acetyl CoA Carboxylase (ACCase) from a diatom in the hopes that there would be an increase in the lipid production. Unfortunately this did not occur.
Primarily two classes of algae became the subject of the research (Figure 2):

Figure 2: Diatom (left) and green algae (right) examples from a sample taken at Sevilleta Nation Wildlife Refuge, Cibola Springs September 2011. Microscope images taken by S. Kintner, September 2011.
In the report A Look Back at the U.S. Department of Energy's Aquatic Species Program: Biodiesel from Algae (Sheehan 1998) section IV.A.2.d. Microalgal Strains for Mass Culture: Source and Genetic Improvements recommendation for future R&D in this field is for a parallel track effort:
These recommendations have been put in place by many researchers, but no research applying algae population studies using metagenomics has been undertaken to date. This is the research proposed with the sampling and testing matrix detailed below based upon this recommendation and the results from bacterial metagenomic research.
4. Metagenomics The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet The National Research Council pulled together the National Institute of Health (NIH), Department of Energy (DOE), National Science Foundation (NSF), Department of Health and Human services and several Universities in the US and United Kingdom to discuss the challenges and functional applications of metagenomics. The study produced the document named “The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet” which will be summarized in this section and related to the proposed study of algal metagenomic samples. Metagenomics applies the power of genomics to the entire community of microbes (algae in this proposal) by eliminating the need to isolate and culture single members. The promise of metagenomics brings within reach the capabilities of microbial communities. Metagenomics harnesses the inherent strengths of genomics, bioinformatics and systems biology and it uniquely analyzes the genomes of many organisms simultaneously while avoiding the pitfall of missing the genomes of non-culturable microbes. Metagenomic brings the power of genomics to study the complex communities where they live. Initially the microbe has been defined as living things not seen by the human eye and microbiologist have often referred to microbes as bacteria. Metagenomics apply not only to bacteria, but also eukaryotes including: fungi, yeast, algae, protozoa and viruses. The study focused primarily on bacteria, but acknowledged metagenomic applications easily extended to other organisms including algae. Eukaryotes have a much larger genome than bacteria housed in a nucleus containing more genes with multiple chromosomes and sequencing the entire genome of eukaryotes presents more challenges than those present in a single bacterial chromosome arrayed with 600k to 12 million base pairs. Characterizing the sampled habitats capitalizes on the algae community metagenomics data. There are many sampling parameters called metadata to define and record such as the quantity (volume), number (organisms in sample), temperature, location (gps coordinates), depth in sampled media, date (season), time of day (day or night) and water/soil chemical composition. The pyrosequencing results paired with the metadata are the tools researchers use to tell the story of the ecological populations. Both genomic and meta data begin to answer the long asked algae population questions of who is there and what are they doing.
5. The Algae Metadata Sampling While sampling the New Mexico algae from a spring the metadata snapshot is taken. An example of a metadata snapshot is shown in Table 1 - Metadata Sampling Matrix,

6. Metagenomic Testing Matrix
Diagram 1: Metagenomic Algae Testing Flow Diagram
Three algae samples are removed from the New Mexico fresh or brine water springs. The samples are immediately put on ice and taken back to the laboratory. A small samples is prepared and shipped to the analytical testing lab where they perform NGS analysis on the samples using a Roche 454 FLEX pyrosequencing. The laboratory sends the data back to understand the significance of the population species diversity.

7. Future Work World energy impacts from the continuously removal of the finite crude oil resource have to be fully realized and the investment in the biofuel infrastructure needs to be put in place now if the future is to continue to run on diesel. Future work on algae populations will need to begin correlating the genomic changes with the real world behavior of population functions, changes and products. The exciting field of study of algae for biofuels has just begun to start to look at a very small but very important eukaryote, algae.
8. Links to Website for Algae Cultivation and Biodiesel
National Renewable Energy Program
National Renewable Energy Program
Aquatic Species Program, NREL
The New Science of Metagenomics
Algae-to-Fuel Research Enjoys Resurgence at NREL
UTEX The Culture Collection of Algae
UTEX Media List
9. References
1. Zhou, Moming and Loder, Asjylyn, (2012) Bloomberg News, Published in TulsaWorld.
2. Sheehan J et al. (1998) A look back at the U.S. Department of Energy's Aquatic Species Program-biodiesel from algae. NREL and US Department of Energy's Office of Fuels Development. NREL/TP-580-24190.
3. Committee on Metagenomics: Challenges and Functional Applications, National Research Council, (2007) The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet, National Academies Press.